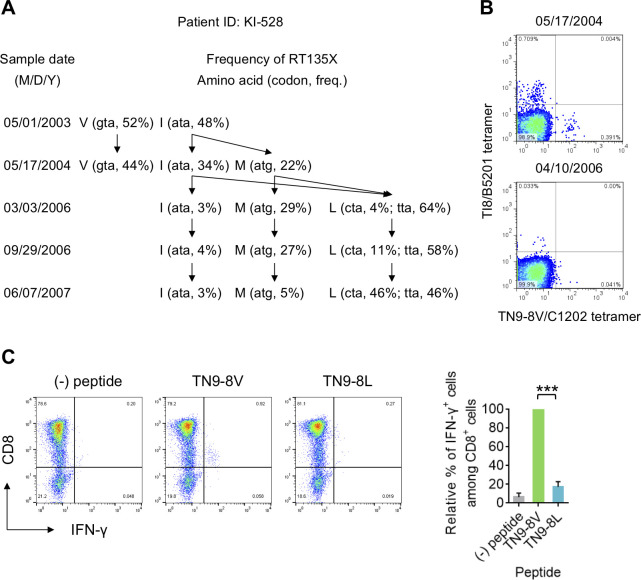
Fig 5. Longitudinal analysis of RT135 mutant selection in an HLA-B*52:01+C*12:02+ individual.
A. Longitudinal sequence analysis at RT135 in an HLA-B*52:01+C*12:02+ hemophiliac individual with HIV-1, KI-528. Frequencies of amino acids at RT135 were determined by both bulk and deep sequencing. Both analyses showed similar results. The amino acids at RT135, their codon usage, and their frequency are shown at each sampling date. Estimated pathways of viral evolution during May 2003-June 2007 are indicated using arrows. B. Detection of TI8- and TN9-8V-specific CD8+ T cells in PBMCs from KI-528 at two different time points. These T cells were detected using TI8/B5201 and TN9-8V/C1202 tetramers. C. Recognition of TN9-8V and TN9-8L peptides by TN9-8V-specific bulk T cells from KI-528. T-cell responses to 721.221-CD4-C1202 cells prepulsed with TN9-8V or TN9-8L peptide were analyzed by ICS, and representative flow cytometric results are shown (left). The relative percent of IFN-γ+ cells among CD8+ cells was calculated by dividing their response frequency to each peptide by their response frequency to TN9-8V (right). Data are shown as the means and SD of triplicate assays. Statistical analysis was conducted using Student's t-test (two-tailed). ***P<0.001.