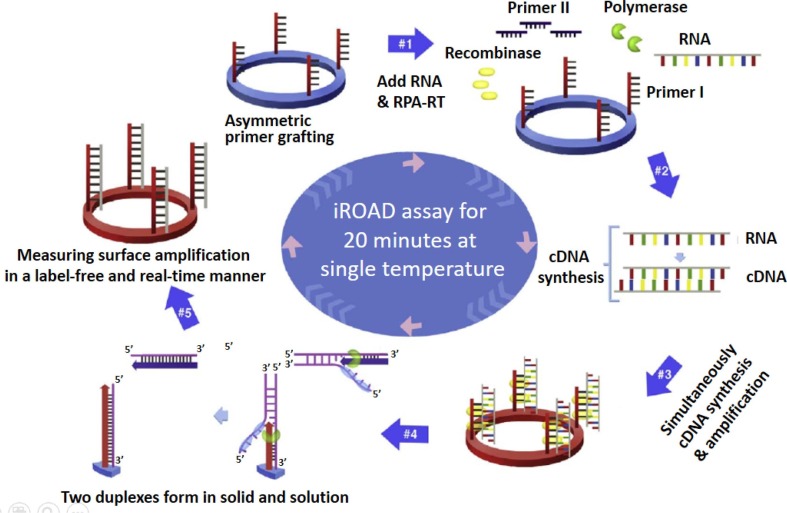
Fig. 3.
Schematic representation of the principle of an isothermal, rapid and label-free one-step RNA amplification/detection (iROAD) assay. First, preparation of the iROAD chip through the primers (forward) grafting on the optical sensor would be needed for a ready-to-use viral RNA detection assay (#1). Then, the mixture containing recombinase polymerase amplification-reverse transcription (RPA-RT) reagents, reverse primers, and extracted RNA is added into the reaction chip (#2). During the isothermal reaction, complementary DNA (cDNA) is synthesized from the RNA template via RNA-RT kit (#3). Thereafter, recombinase/primer complexes bind to double-stranded target cDNA and facilitate strand exchange at a constant temperature. After the displaced strand forms a D-loop by gp32 (sky blue), the immobilized primers are extended by polymerase (light green) on the surface of the silicon microring resonator (#4). The formation of two duplexes is caused by the amplification of the solid and the solution. The exponential RNA amplification after the reverse transcription based on the asymmetric assay is achieved by the repletion of the process (#5). The amplification and detection of the target is simultaneously monitored by measuring the wavelength shift on an optical sensor for 20 min. Figures modified from Ref. [78] and used with permission.