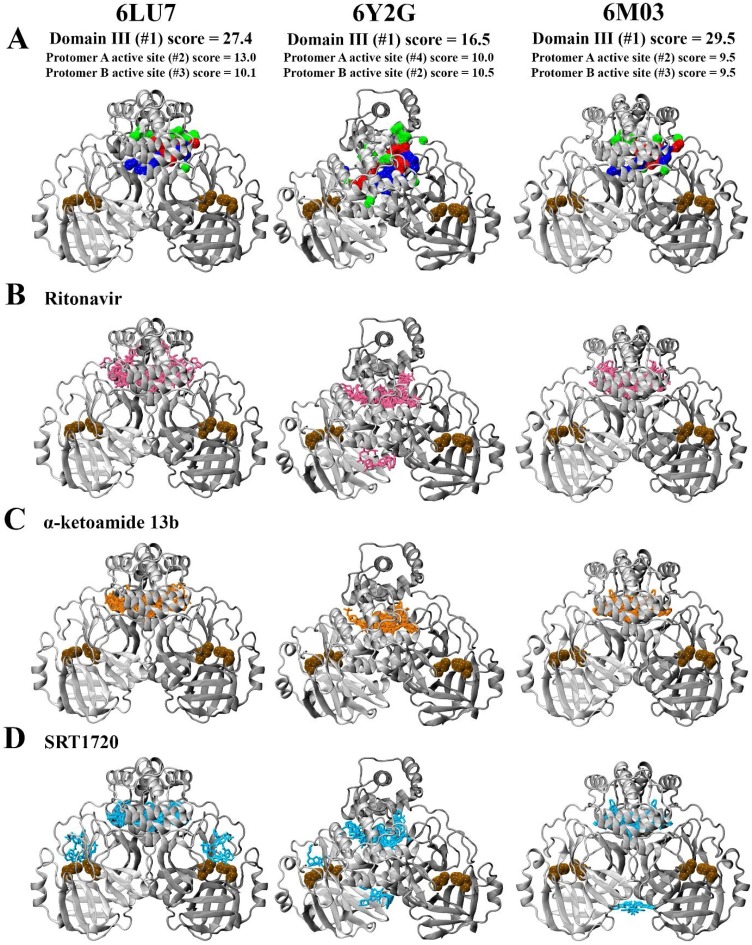
Fig. 6.
Analysis of binding sites in structures of the SARS-CoV-2 Mpro (PDB ID: 6LU7, 6Y2G, and 6M03) using the PrankWeb Server A). The most likely binding pocket indicated by pocket score is depicted, highlighted in surface representation with non-polar residues in white, basic residues in blue, acidic residues in red, and polar residues in green. Blind docking of B) ritonavir, C) α-ketoamide 13b, and D) SRT1720 to the protease dimer is shown. Catalytic dyad residues His41 and Cys145 are indicated in ochre to denote the location of the active site.