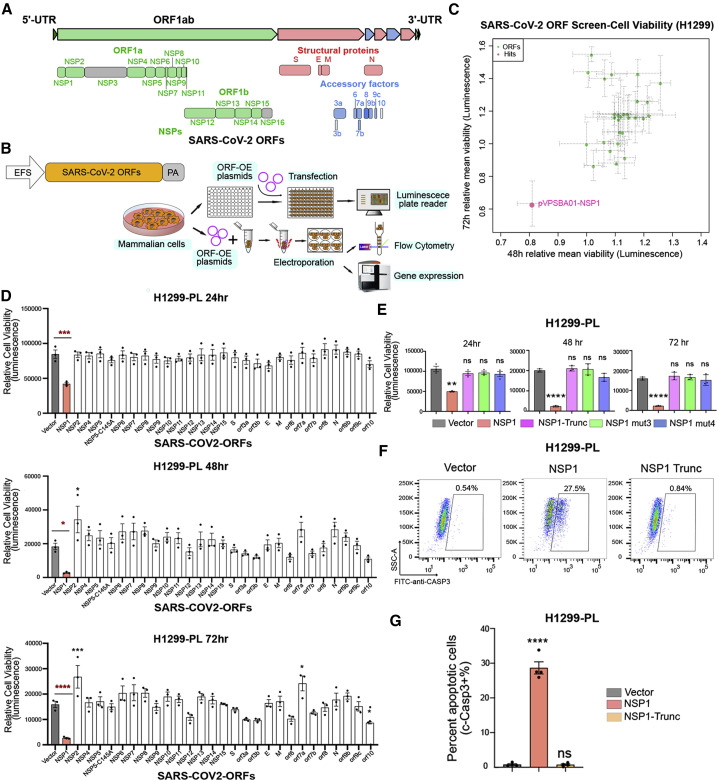
Figure 1.
SARS-CoV-2 ORF Mini-screen Identified Nsp1 as a Key Viral Protein with Host Cell Viability Effect
(A) Schematics of viral protein coding frames along SARS-CoV-2 genome. Colored ORFs indicate the ones used in this study, while two ORFs in gray are not (Nsp3 and Nsp16).
(B) Schematics of molecular and cellular experiments of viral proteins.
(C) Scatterplot of SARS-CoV-2 ORF mini-screen for host viability effect in H1299 cells, at 48 and 72 h post-ORF introduction. Each dot represents the mean normalized relative viability of host cells transfected with a viral protein encoding ORF. Dashed-line error bars indicate SDs (n = 3 replicates). Pink color indicates hits with p < 0.05 (one-way ANOVA, with multiple-group comparison).
(D) Bar plot of firefly luciferase reporter measurement of viability effects of SARS-CoV-2 ORFs in H1299-PL cells, at 24, 48, and 72 h post-ORF introduction (n = 3 replicates).
(E) Bar plot of firefly luciferase reporter measurement of viability effects of Nsp1 and three Nsp1 mutants (truncation, mut3: R124S/K125E and mut4: N128S/K129E) in H1299-PL cells, at 24, 48, and 72 h post-ORF introduction (left, middle, and right panels, respectively) (n = 3 replicates).
(F) Flow cytometry plots of apoptosis analysis of Nsp1 and loss-of-function truncation mutant in H1299-PL cells, at 48 h post-ORF introduction. Percentage of apoptotic cells was gated as cleaved caspase-3-positive cells.
(G) Quantification of flow-based apoptosis analysis of Nsp1 and loss-of-function truncation mutant in H1299-PL cells, at 48 h post-ORF introduction.
For all bar plots in this figure, bar height represents mean value and error bars indicate standard error of the mean (SEM) (n = 3 replicates for each group). Statistical significance was accessed using ordinary one-way ANOVA, with multiple-group comparisons where each group was compared with empty vector control, with p values subjected to multiple-testing correction by FDR method (ns, not significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001). See also Figure S1.