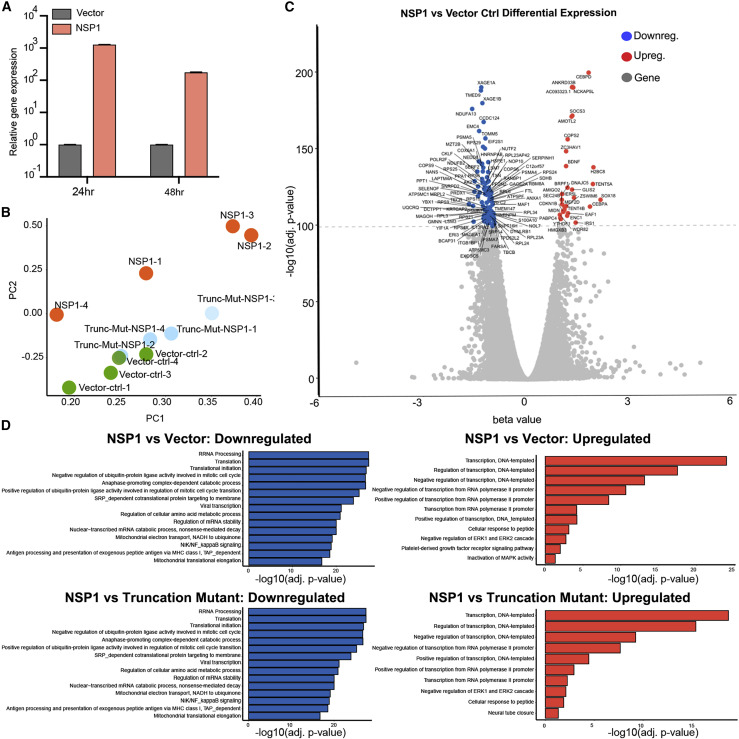
Figure 2.
Transcriptome Profiling of H1299 Cells Introduced with NSP1 and NSP1 Truncation Mutant by RNA-Seq
(A) Quantitative PCR (qPCR) confirmation of NSP1 overexpression, at 24 and 48 h post-electroporation (n = 3 replicates).
(B) Principal-component analysis (PCA) plot of the entire mRNA-seq dataset, showing separation between Nsp1, vector control, and Nsp1 truncation mutant groups, all electroporated into H1299-PL cells and harvested 24 h post-electroporation. RNA samples were collected as quadruplicates (n = 4 each group).
(C) Volcano plot of differential expression between of Nsp1 versus vector control electroporated cells. Top differentially expressed genes (FDR-adjusted q < 1e-100) are shown with gene names. Upregulated genes are shown in orange. Downregulated genes are shown in blue.
(D) Bar plot of top enriched pathway analysis by DAVID biological processes (BP). Nsp1 versus vector control (top) or Nsp1 versus Nsp1 mutant (bottom); highly downregulated (left) and upregulated (right) genes are shown (q < 1e-30).
See also Figure S2.