Figure 1.
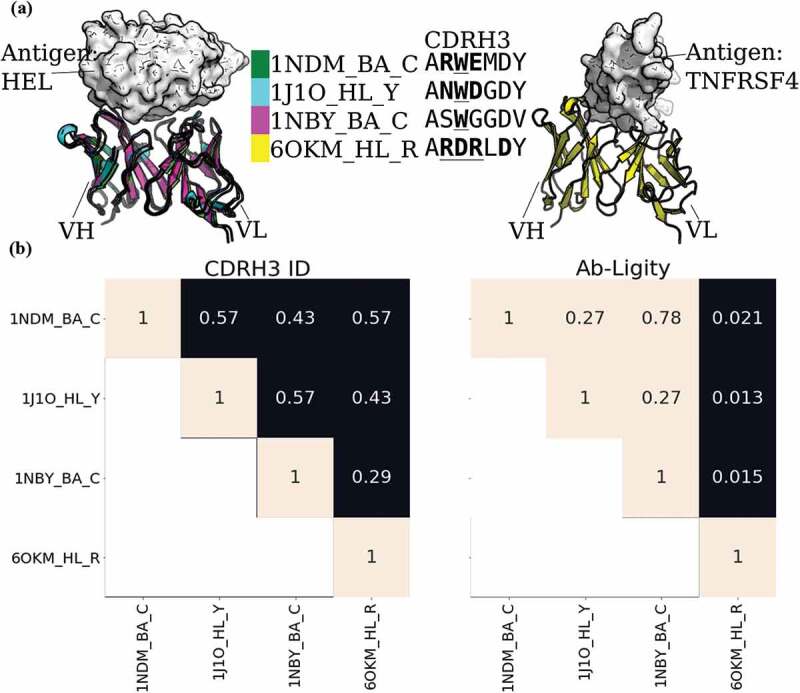
Analysis of anti-lysozyme (HEL) antibodies with dissimilar CDRH3 sequences and highly similar epitopes
(A) Structural superposition of three anti-lysozyme and one anti-TNFRSF4 antibodies co-crystallized with their antigens (lysozyme or TNFRSF4) in white. The three anti-lysozyme antibodies were HyHEL-26 (1NDM_BA_C), HyHEL-10 L-Y50F mutant (1J1O_HL_Y) and HyHEL-63 (1NBY_BA_C); and the anti-TNFRSF4 antibody is 3C8 (6OKM_HL_R). The antigens from the three anti-lysozyme antibody crystal structures are aligned. The legend shows the colors of the antibodies with their PDB codes followed by the heavy-light chain and antigen chain identifiers, separated by (‘_’). The CDRH3 sequences are displayed next to the respective antibody identifiers. Crystal paratope residues within the CDRs are in bold, and Parapred-predicted paratopes within the CDRs are underlined. The other five CDR sequences are listed in Supplementary Table S15. (B) Heatmaps of CDRH3 sequence identity and Ab-Ligity paratope similarity. The row and column labels correspond to the structures shown in (A). Ab-Ligity paratope similarity is calculated on the antibody model and predicted paratope as outlined in the Methods section. Pairs of antibodies with CDRH3 identity of >0.80 would have been considered similar by sequence-based metric. For Ab-Ligity, a similarity score of >0.1 suggests that the antibodies bind to highly similar epitopes.
