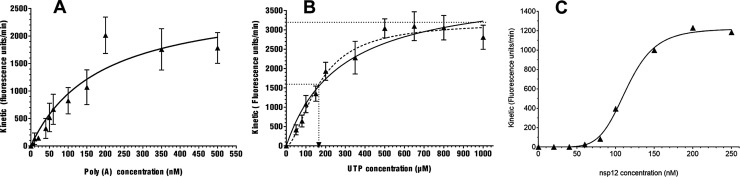
Fig. 1.
Setting up of the SARS-CoV-RTC activity experimental conditions based on a fluorescent readout.
(A)Poly(A) template variation with SARS nsp12 in complex with nsp7L8.
Velocity values of SARS nsp12 (150 nM) in complex with nsp7L8 (1.5 μM) was measured for various concentrations in Poly (A) template (5; 10; 20; 40; 60; 80; 100; 150; 200; 350; 500 nM). Using the Prism software, velocity values of each condition were determined by calculating the slope of the linear phase of the kinetic and then plotted against the Poly(A) template concentration by using Michaelis-Menten fitting. Data were the results of three independent experiments.
(B) Nucleotide variation with SARS nsp12 in complex with nsp7L8.
Velocity values of SARS nsp12 (150 nM) in complex with nsp7L8 (1.5 μM) was measured for various concentrations in UTP (50;80;100;150;200;350;500 ;650;800 and 1000 nM). Using the Prism software, velocity values of each condition were determined by calculating the slope of the linear phase of the kinetic and then plotted against the UTP concentration by using Michaelis-Menten equation (full line) or Hill equation (dot line) to determine the apparent Km (UTP) of the SARS-CoV-RTC. Data were the results of three independent experiments.
(C) Variation in SARS nsp12 in complex with nsp7L8.
Velocity values obtained during a time course with 350 nM Poly (A) and 500 μM UTP were plotted against different concentrations in SARS nsp12 in complex with nsp7L8. The concentration ratio nsp12:nsp7L8 was conserved at 1:10. Curve was fitted according the Hill equation.