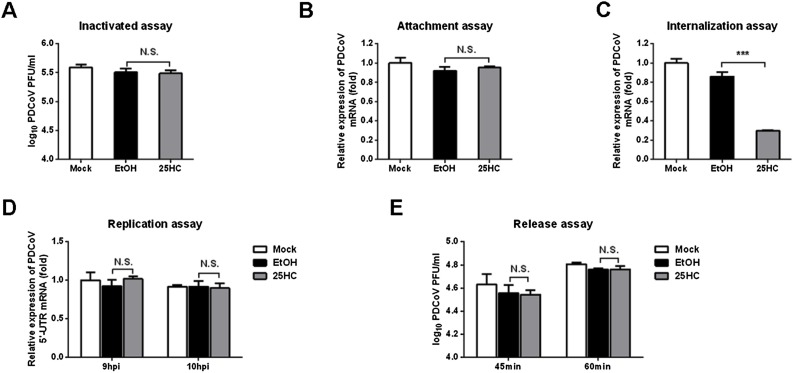
Fig. 5.
25HC inhibits PDCoV invasion. (A) Viral inactivation assay. PDCoV and 25HC were incubated at 37 °C for 3 h. LLC-PK1 cells cultured in six-well plates were prechilled at 4 °C for 1 h. The growth medium was replaced with a mixture of 25HC (12.5 μM) or EtOH and PDCoV. After incubation at 4 °C for another 2 h, the cells were washed with precooled PBS, covered with overlay medium, incubated at 37 °C for a further 1–2 days, and assessed using a plaque assay. (B) Adsorption assay. IPI-FX cells cultured in 24-well plates were pretreated with 25HC (12.5 μM) or EtOH for 1 h at 37 °C and then prechilled at 4 °C for 1 h. The growth medium was then replaced with a mixture of 25HC (10 μM) or EtOH and PDCoV (MOI 0.5) in the presence of 2.5 μg/mL trypsin for 1 h at 4 °C. After washing three times with prechilled PBS, PDCoV N protein mRNA levels were measured by qRT-PCR. (C) Penetration assay. IPI-FX cells cultured in 24-well plates, prechilled at 4 °C for 1 h and then infected with PDCoV (MOI 0.5) in the presence of 2.5 μg/mL trypsin for 1 h at 4 °C. The virus-containing medium was replaced with fresh medium containing 25HC (12.5 μM) or EtOH. The temperature was raised to 37 °C for 3 h and then the cells were washed with PBS, pH 3. PDCoV N protein mRNA levels were assessed by qRT-PCR. (D) Replication assay. IPI-FX cells were infected with PDCoV (MOI 0.5) in the presence of 2.5 μg/mL trypsin at 37 °C for 8 h. Cell-free virus particles were removed. Cells were treated with fresh medium containing 25HC (12.5 μM) or EtOH for 9 or 10 h. The cells were collected for qRT-PCR to assess levels of negative-sense PDCoV RNA. (E) Release assay. IPI-FX cells were inoculated with PDCoV (MOI 0.5) at 4 °C for 1 h. At 12 hpi, the inoculum was replaced with fresh medium containing 25HC (12.5 μM). Cell supernatants were harvested at 45 and 60 min and titrated using a plaque assay. All data are presented as the means ± standard deviations of three independent experiments. *P < 0.05 and **P < 0.01.