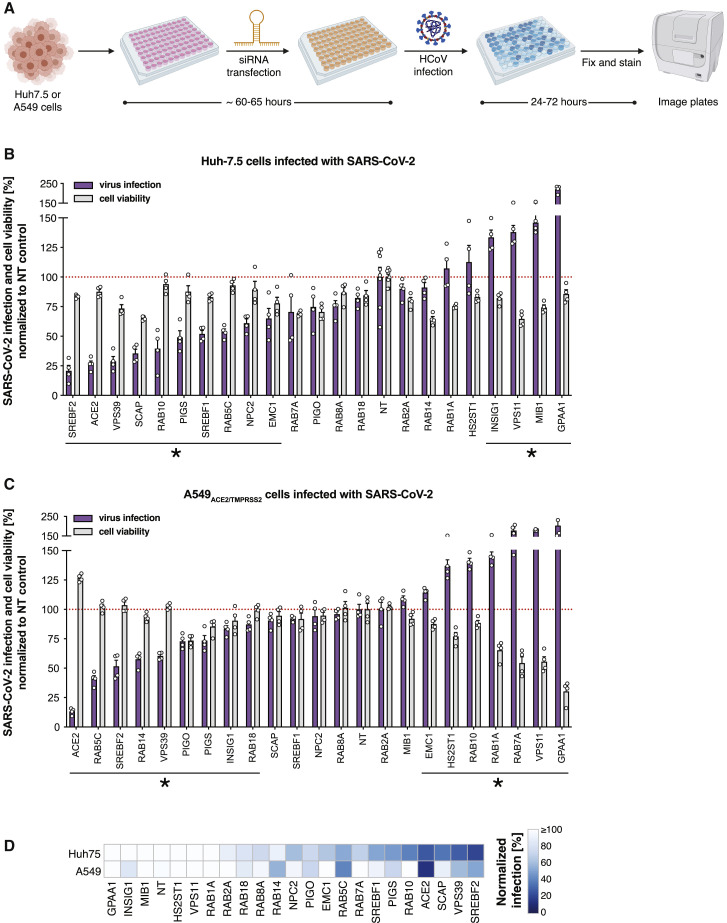
Figure 3.
Validation of high-confidence SARS-CoV-2 protein interactome factors during infection by SARS-CoV-2
(A) Experimental scheme for candidate validation by using arrayed siRNA knockdown. Diagram created with BioRender.com.
(B) Candidate validation in Huh-7.5 cells at 37°C using 5,000 SARS-CoV-2 plaque-forming units (PFU) per well. Plates were quantified at 24 h after infection. Data represent the average ± SEM of n = 4 replicates per target gene. Significance for viral infection percentage values was evaluated by using Student's t test (2-tailed, unpaired) as compared with those of non-targeting (NT) controls (set at 100%). ∗ p < 0.05.
(C) Candidate validation in A549ACE2/TMPRSS2 cells at 37°C using 2,500 SARS-CoV-2 PFU per well. Plates were quantified at 24 h after infection. Data represent the average ± SEM of n = 4 replicates per target gene. Significance for viral infection percentage values was evaluated by using Student's t test (2-tailed, unpaired) as compared with those of non-targeting (NT) controls (set at 100%). ∗p < 0.05.
(D) Heatmap representation of data from (B) and (C).
See also Figures S2 and S3.