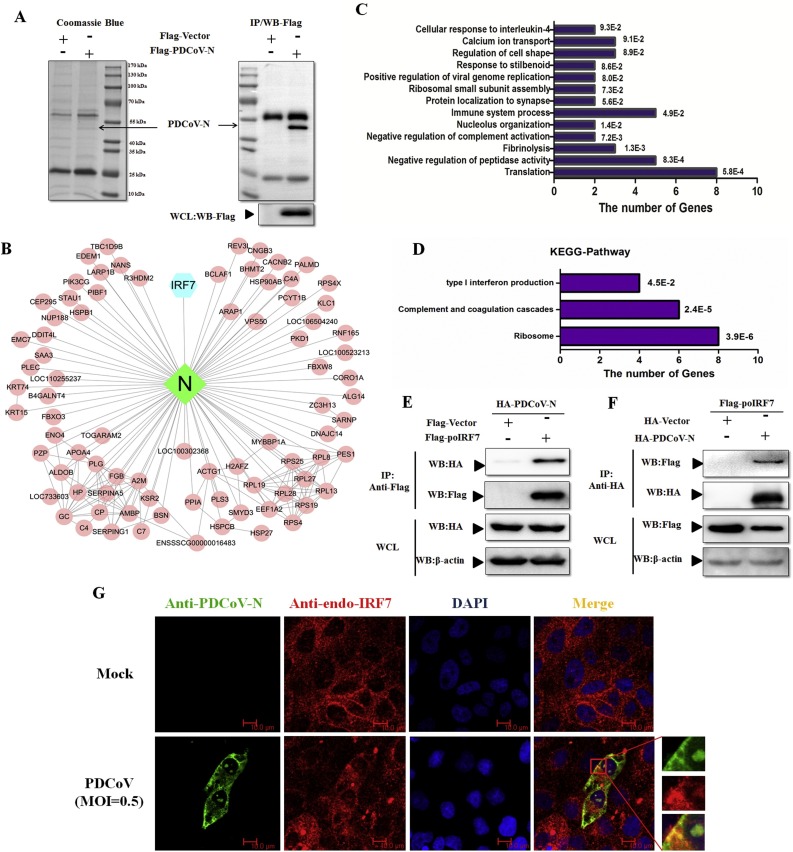
Fig. 2.
PDCoV N protein interacts with poIRF7. (A) The immunoprecipitates of PDCoV N protein in PK-15 cells were separated with SDS-PAGE, and then detected by Coomassie blue staining or Western blot with anti-Flag antibody. (B) The interaction net-work was built with Cytoscape software. (C, D) Results of analysis of (C) Gene ontology (GO) enrichments and (D) the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichments of the specific proteins identified in the Flag-PDCoV N immunoprecipitates. The Y-axis is the name of each category, while the X-axis is the number of genes enriched in each category. Their p values were shown at the end of each bar. (E, F) HEK293 T cells were co-transfected with Flag-poIRF7 or Flag-Vector and HA-PDCoV-N or HA-Vector expression plasmids for 28 h. Cells were lysed for co-immunoprecipitation (Co-IP) assay with anti-Flag (IP: anti-Flag) or anti-HA (IP: anti-HA) affinity gel. The whole cell lysates (WCLs) and immunoprecipitants were detected by Western blot with anti-Flag, anti-HA or anti-β-action antibody. (G) The confocal microscope assay was used to detect the localization of PDCoV-N (green) and endogenous poIRF7 (endo-poIRF7, red) in the mock or PDCoV infected LLC-PK1 cells, respectively. DAPI (blue) stained the cell nuclei. Fluorescent images were acquired with a confocal laser scanning microscope (scar bar: 10 μm).