Figure 1.
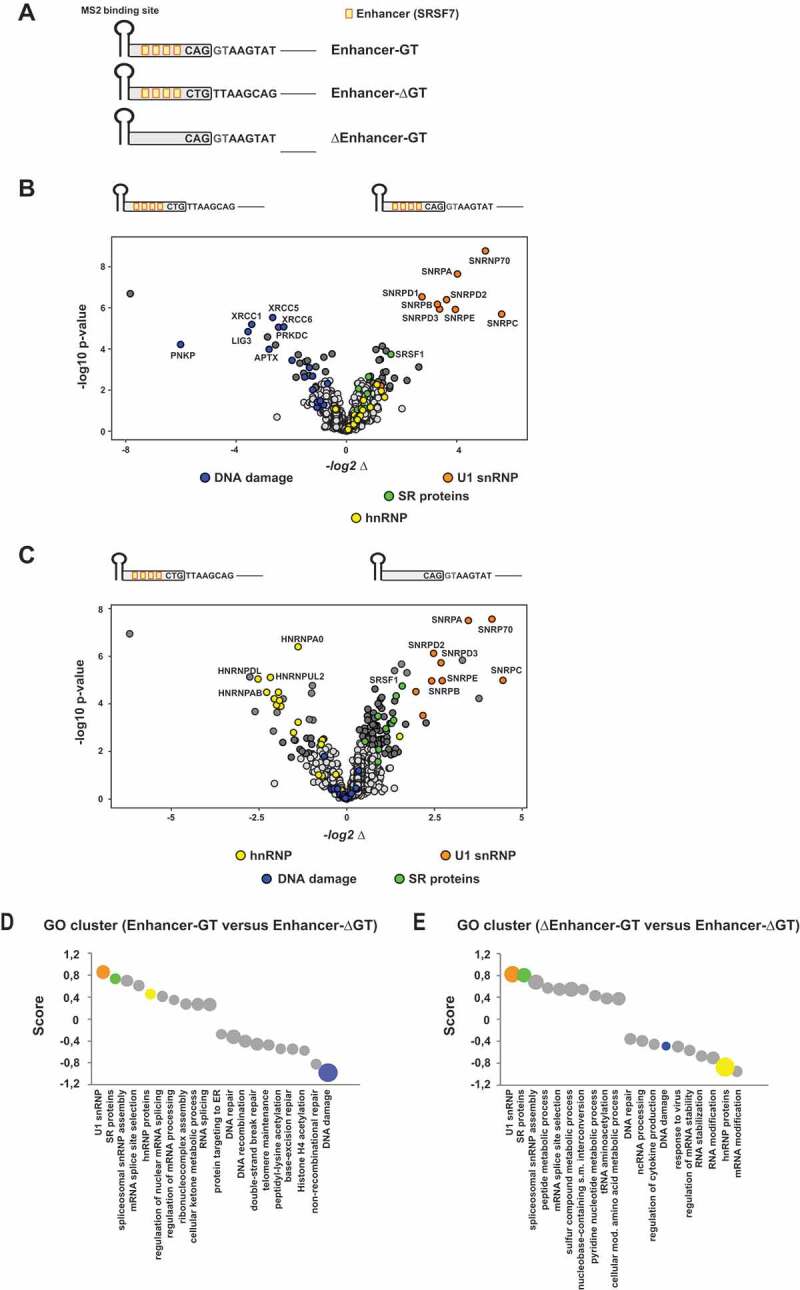
Profiling of the functional splice siteinteractome
(A) Schematic of the RNA substrates used for RNA in vitro pulldown assays. All in vitro transcribed RNAs contained an MS2 binding site at the 5ʹ-end that was recognized by recombinant MS2 coat protein (added to the nuclear extracts) and served as a precipitation control. RNAs were covalently coupled to agarose beads and bound fractions subsequently analysed by mass spectrometry (MS). Orange boxes indicate single splicing enhancer sequences that had been demonstrated before to efficiently enhance splice site activation [15,23,62]. (B, C) MS analysis identified the functional 5’ss (enhancer plus GT 5’ss) interactome (U1 snRNP proteins are highlighted in orange). Absence of the 5’ss sequence strongly reduced the levels of precipitated U1 snRNP proteins (B-C). In addition, the enhancer sequence revealed enriched binding for hnRNP proteins (C) (highlighted in green). Volcano Plots had been generated using InstantClue (www.instantclue.uni-koeln.de). (D, E) One dimensional Gene Ontology (GO) enrichment analysis was performed using Perseus. The top 10 non-redundant categories showing a shift to higher or lower abundances are presented. The size of the bubbles indicates the associated – log10 p-values. Ranges for – log10 p-values are 3.4 (smallest bubble) to 11.4 (largest Bubble) in E, 2–11.3 in F and 2.1–9.6 in G. The U1 snRNP complex annotation cluster is highlighted in orange, DNA repair cluster in blue and hnRNP cluster in yellow.
