Figure 5.
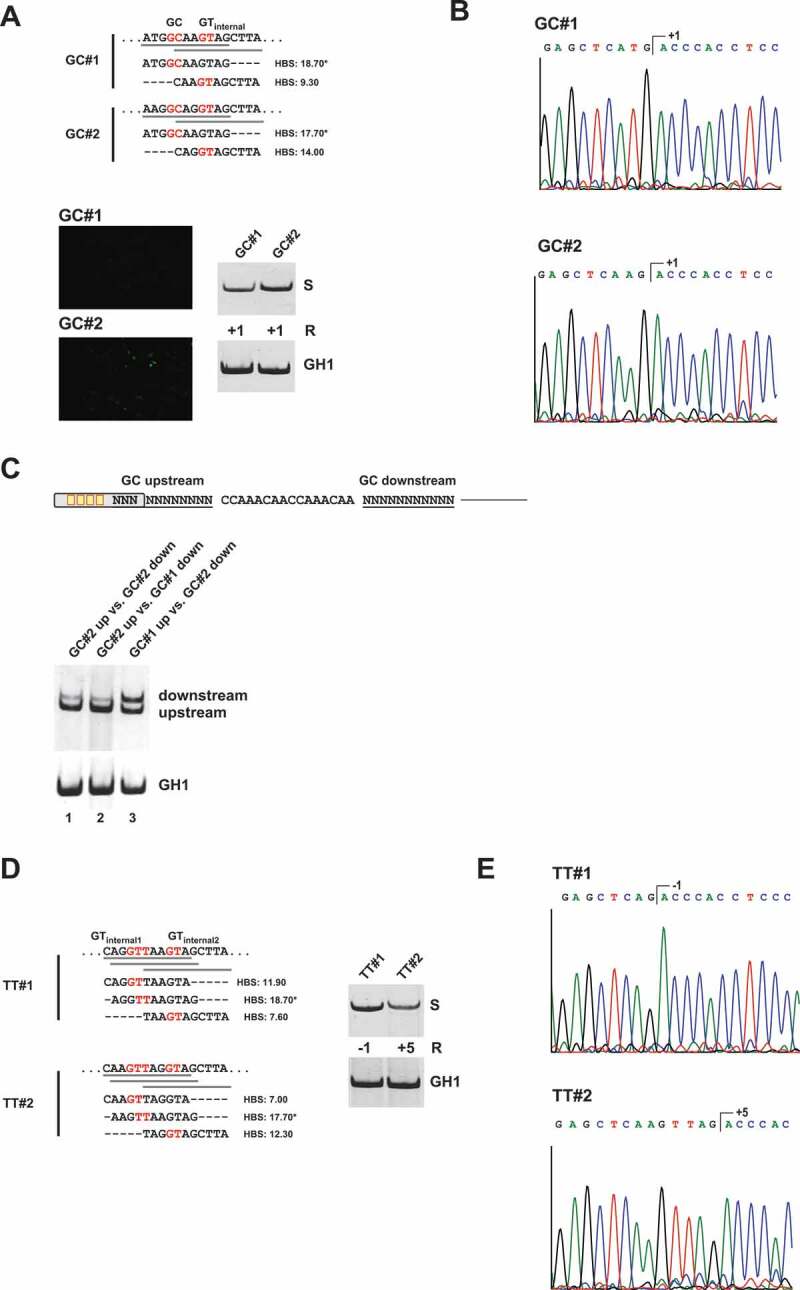
A noncanonical GC splice site benefits from presence of an internal GT site at position +5/+6
(A) Schematic of the HIV-1-based SV-env/eGFP splicing reporter containing different GC splice sites (on top). Sequence variations and H-Bond scores of the GC splice sites and the internal GT sequences at position +5/+6 are shown below. Splicing-dependent eGFP expression was monitored by fluorescence microscopy (see below). RT-PCR analyses of spliced reporter mRNAs were performed as described before. Splicing positions (R) are indicated below. (B) Sequencing results of GC splice site usage shown in (A). Polyacrylamide (PAA) bands were isolated, reamplified with primer pair #3210/#3211 and sent to sequencing analysis using primer #3210. (C) RT-PCR analysis of competing GC splice sites. (D, E) RT-PCR analysis and sequencing results of TT splice sites with variations of the internal GT sequence at position −1/+1 or +5/+6. *H-Bond score had been calculated by inserting a GT at position +1/+2; S: spliced; +1: cleavage at position +1; −1: cleavage at positions −1; +5: cleavage at position +5.
