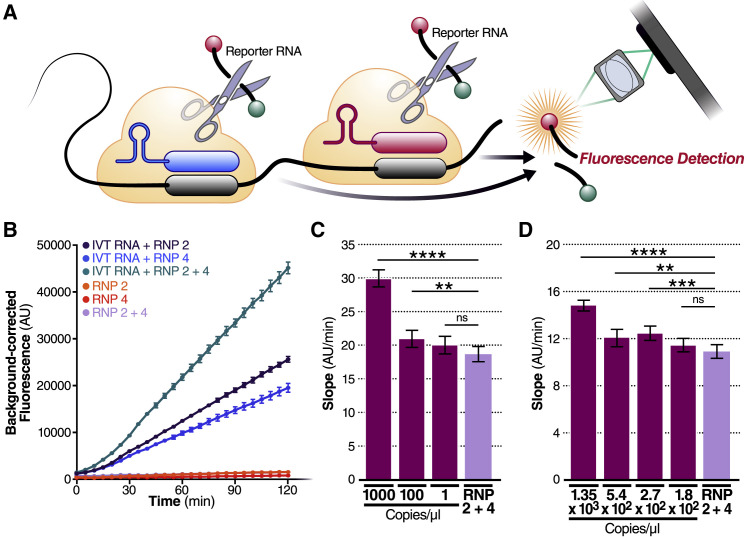
Figure 2.
Combining crRNAs improves sensitivity of Cas13a
(A) Schematic of two different RNPs binding to different locations of the same SARS-CoV-2 RNA, leading to cleavage of the RNA reporter and increased fluorescence.
(B) RNPs made with crRNA 2 and crRNA 4 individually and in combination (50 nM total RNP concentration per reaction) were tested against 2.89 × 105 copies/μL (480 fM) of SARS-CoV-2 IVT N gene RNA and compared to fluorescence from no target RNA RNP alone controls (“RNP 2,” “RNP 4,” and “RNP 2+4”). Background correction of fluorescence was performed by subtraction of reporter-alone fluorescence values. Data are represented as mean ± standard error of the difference between means of three technical replicates.
(C) Limit of detection of crRNA 2 and crRNA 4 in combination was determined by combining 50 nM of RNP 2 and 50 nM of RNP 4 (100 nM total) against 1,000, 100, and 1 copy/μL of SARS-CoV-2 IVT RNA (n = 3, technical replicates). Slope of the curve over 2 h was calculated by performing simple linear regression of data merged from replicates and is shown as slope ± 95% confidence interval. Slopes were compared to the no target RNA RNP alone control using ANCOVA: ∗∗∗∗p < 0.0001, ∗∗p = 0.0076, ns = not significant.
(D) Limit of detection of crRNA 2 and crRNA 4 in combination was determined by combining 50 nM of RNP 2 and 50 nM of RNP 4 (100 nM total) against 1.35 × 103, 5.4 × 102, 2.7 × 102, and 1.8 × 102 copies/μL of genomic SARS-CoV-2 viral RNA as quantified by qPCR (n = 3, technical replicates). Slope of the curve over 2 h was calculated by performing simple linear regression of data merged from replicates and is shown as slope ± 95% confidence interval. Slopes were compared to the no target RNA RNP alone control using ANCOVA: ∗∗∗∗p < 0.0001, ∗∗∗p = 0.0002, ∗∗p = 0.0023, ns = not significant.