Figure S1.
Individual crRNAs quantitatively detect SARS-CoV-2 RNA, related to Figure 1
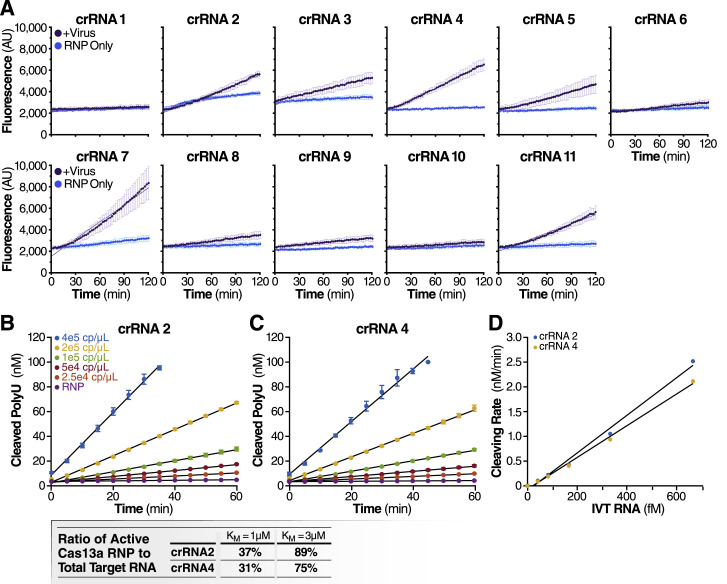
(A) Cas13a RNPs made individually with each N gene crRNA (final RNP complex concentration of 100 nM) were tested against extracted SARS-CoV-2 viral RNA. Background fluorescence by the individual RNP in the absence of target RNA is shown as “RNP Only.” Raw fluorescence values over 2 h is shown. Data are represented as mean ± SD of three technical replicates.
(B and C) Cas13a reaction rate is linearly proportional to the target RNA concentration.
(B) The reaction rate of Cas13a was measured by adding a range of concentrations of IVT N gene RNA to reactions that contain 100 nM of Cas13a RNP with crRNA 2 and 400 nM of polyU reporter (error bars indicate the standard deviation of triplicate measurements). The reaction rate is determined by fitting a linear curve to the data (black curves).
(C) The reaction rate of Cas13a for a range of IVT N gene RNA concentrations as measured in Figure S1B but with crRNA 4 used in place of crRNA 2.
(D) Cas13a reaction rate – either with crRNA 2 or crRNA 4 – scales linearly with the target IVT RNA concentration (R2 = 0.990 for crRNA 2 and 0.996 for crRNA 4). Assuming that Cas13a enzymatic activity can be described by the Michaelis-Menten kinetics model, and that the amount of IVT RNA sets the upper limit of active Cas13a RNP, we predict the ratio of active Cas13a to IVT RNA in for Kcat = 600/s and KM = 1 μM or 3 μM, which is the range of KM previously found for Cas13b (Slaymaker et al., 2019).