Figure 3.
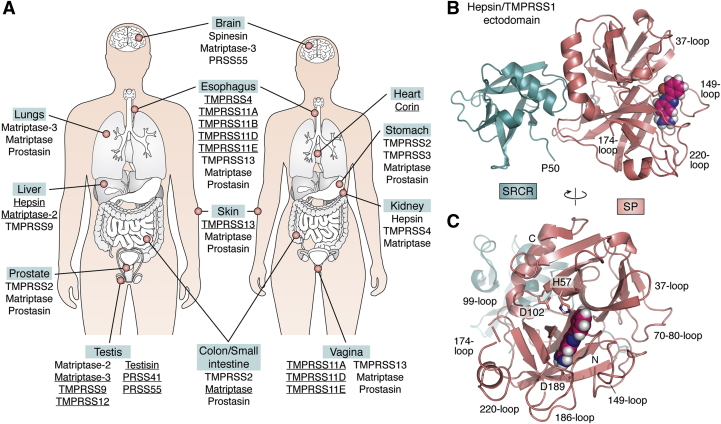
Expression patterns and structure of membrane-associated serine proteinases.A, organs with highest expression levels of specific MASPs. Data for some organs are indicated only for males or females, although there are no known sex-associated differences in MASP expression in these organs. Data from the GTEx Portal (gtexportal.org) (see also Fig. 4A). B–C, 3D crystal structure of human hepsin/TMPRSS1 ectodomain, represented as a cartoon highlighting major secondary structure elements; loops that shape the active-site cleft are noted. (After PDB entry 1P57: N-terminal SRCR module (deep-teal cyan); serine proteinase domain (deep salmon-red)). A small-molecule inhibitor bound in the S1 specificity pocket of the serine proteinase domain (2-{5-[amino(iminio)methyl]-1H-benzimidazol-2-YL}benzenolate) is shown as color-coded spheres (carbon, pink; oxygen, red; nitrogen, blue; and hydrogen, gray). B, side view, highlighting the proximity of the N-terminal residue of the SRCR domain, Pro50, to the transmembrane helix, Gly24-Leu44. This locates also the rigidly attached catalytic domain of the proteinase essentially flat against the cell membrane (74). A similar localization should be expected for the catalytic domains of other MASPs, which implies that the Arg815-Ser816 bond in the S protein would be cleaved close to the cell membrane, facilitating rapid interaction with the exposed viral fusion peptide and escape from immune surveillance. C, view of the proteinase in the “standard orientation”, e.g., with active-site residues (given with all their nonhydrogen atoms, color-coded) facing the viewer and substrates running from left to right. The N and C termini of the catalytic chain are noted (Ile16 and Thr253, respectively) as well as Asp189 at the bottom of the S1 pocket, which is largely responsible for the recognition and cleavage of substrates after a basic Arg/Lys residue.