Figure 1.
Sequence similarity between SARS-CoV and SARS-CoV-2
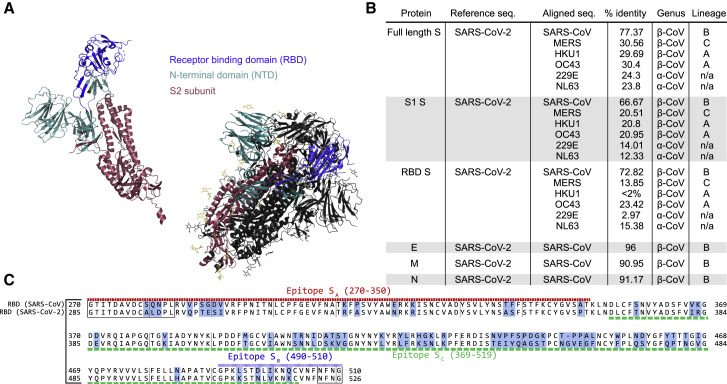
(A) Similarity scores for each of the SARS-CoV-2 structural proteins compared with SARS-CoV and other common coronaviruses. Similarity in the S protein is substantially lower than for the other structural proteins.
(B) Sequence alignment of the receptor binding domain (RBD) of SARS-CoV and SARS-CoV-2. Regions of difference are highlighted in blue, while the epitopes of the antibodies used in this study are underlined according to their designations in Table 1. The boxed regions fall outside of the canonical RBD sequence but are included because of overlap with the above epitope regions.
(C) Crystal structure of the S protein color coded by domain both as monomer and in the functional homotrimeric form in which one of the monomers is colored, while the other two are shown in white. As shown, the NTD and RBD compose the majority of the S1 region.
See also Figure S1.