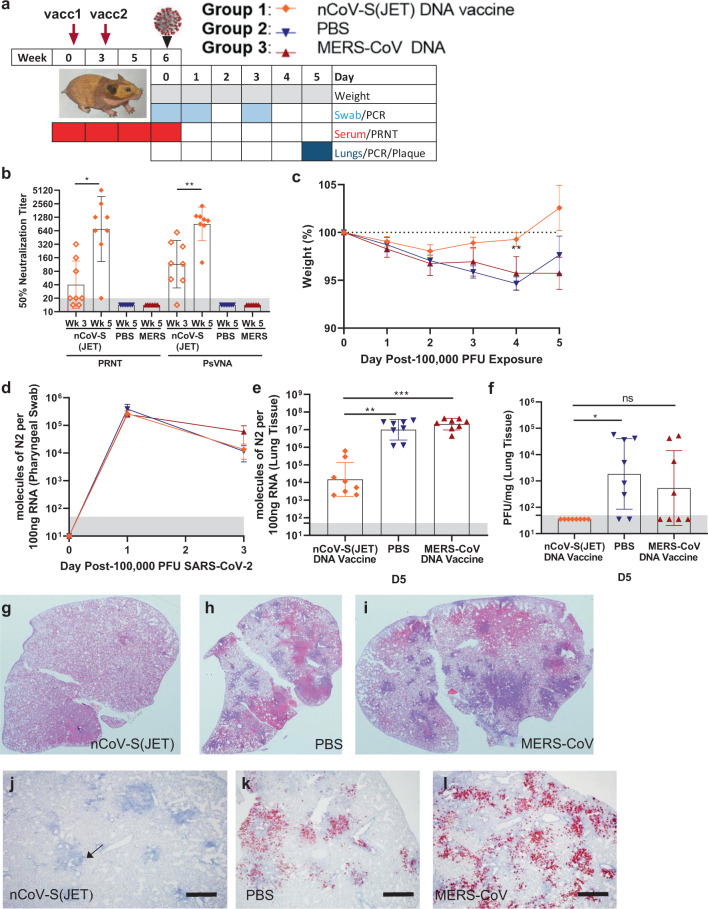
Fig. 1. Evaluation of nCoV-S(JET) DNA vaccine in Syrian hamsters.
a Experimental design. Groups of 8 hamsters each were vaccinated (vacc) with the nCoV-S(JET) DNA vaccine, PBS, or a MERS-CoV DNA vaccine and then challenged with 100,000 PFU of SARS-CoV-2 virus by the intranasal route. b PRNT50 and PsVNA50 titers from serum collected at indicated timepoints after 1 (open symbols) and 2 (closed symbols) vaccinations (assay limit = 20, gray shade). Bars represent GMT ± SD. c Average animal weights relative to starting weight. Symbols represent mean ± SEM. Viral RNA in d pharyngeal swabs and e lung homogenates (assay limit = 50 copies, gray shade). Bars GMT ± SD. f Infectious virus as measured by plaque assay (assay limit = 50 PFU, gray shade). Bars GMT ± SD. Bright field imagery of H&E staining of lung sections from g nCoV-S(JET) DNA, h PBS, or i MERS-CoV vaccinated hamsters where purple indicates areas of consolidation. ISH to detect SARS-CoV-2 genomic RNA in lung sections of j nCoV-S(JET) DNA, k PBS, and l MERS-CoV vaccinated hamsters. Rare, positive labeling in nCoV-S(JET) DNA vaccinated hamster lung sections were detected (arrows). Asterisks indicate that results were statistically significant, as follows: *p < 0.05; **p < 0.01; ***p < 0.001; ns not significant. Scale bars = 400 microns. Hamster drawing was provided by Jake Hooper (hjake@vt.edu), with permission.