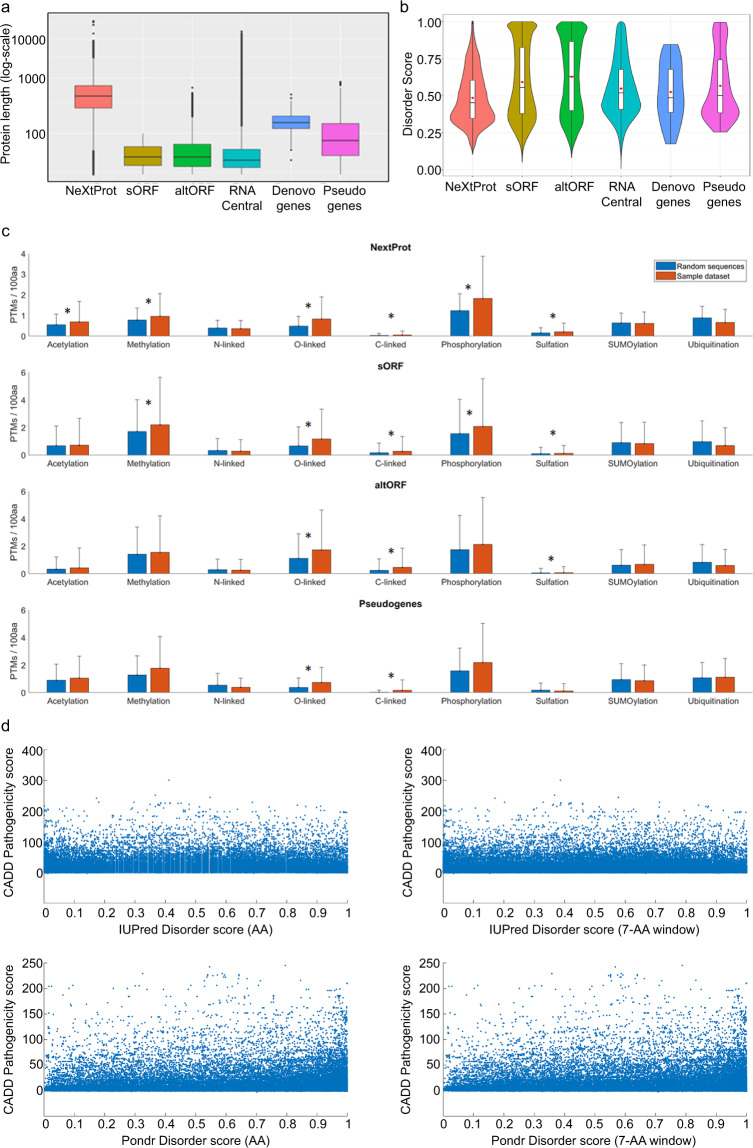
Fig. 4. Novel proteins are smaller in length and show increased structural disorder but still have regulatory regions.
a Amino acid length distribution of known human proteins from NeXtProt, and potential novel proteins encoded by nORFs: sORFs, altORFs, Pseudogenes, Denovogenes, and all possible translated amino acid sequences from RNAcentral. b Average disorder scores of proteins in NeXtProt compared to average disorder scores of proteins encoded by nORFs, predicted by PONDR. c PTM sites in protein sequences were predicted using the ModPred tool. The predicted densities of nine PTM modifications for each dataset (NeXtProt, sORFs, altORFs, or pseudogenes) were compared against the predicted PTM densities in individual control datasets (random AA sequences generated to have the same average amino acid composition and length distribution as the original dataset). d Disorder scores were computed at either amino-acid resolution, or for a 7-AA window around the mutated residue. The analysis did not reveal any correlations between CADD scores and predicted disorder scores.