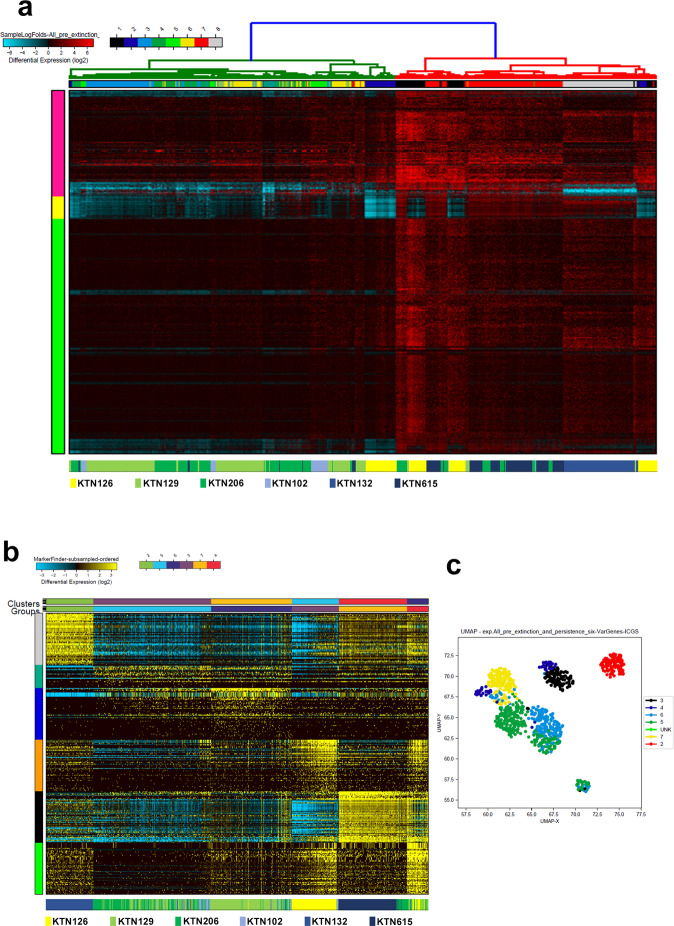
Fig. 1. Iterative Clustering and Guide-gene Selection (ICGS) of TNBC-derived single cells pre neoadjuvant therapy.
a Hierarchical clustering of 872 single cells derived from three extinction (KTN126, KTN129, and KTN206) and three persistence (KTN102, KTN132, and KTN615) triple-negative breast cancer (TNBC) prior to neoadjuvant chemotherapy based on lncRNA transcriptome. b Unsupervised single-cell population identification using ICGS algorithm conducted on 577 single cells derived from the extinction (KTN126, KTN129, and KTN206) and 295 cells derived from the persistence (KTN102, KTN132, and KTN615) group prior to neoadjuvant treatment. Data are presented as heat map with the corresponding single-cell cluster on top. Color scale displays differential gene expression (log2). The lower legend indicated the source of each cell. c Uniform manifold approximation and projection (UMAP) dimensionality reduction analysis of 872 single cells from the same cohort prior to neoadjuvant treatment.