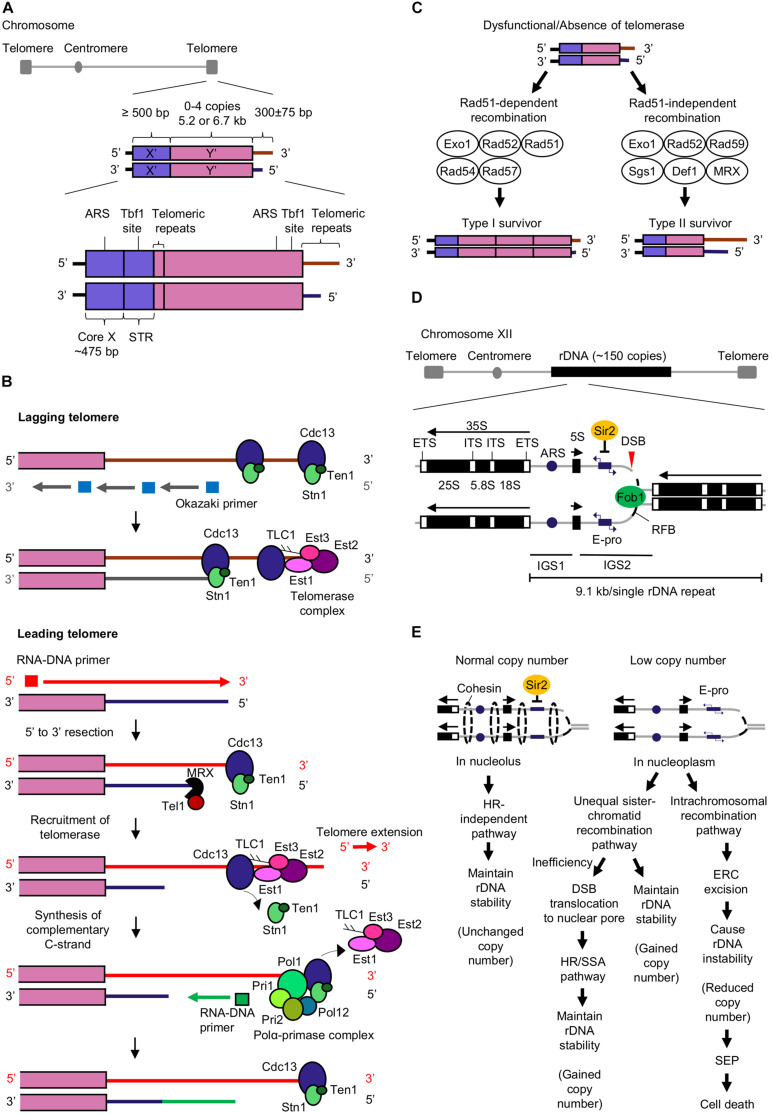
FIGURE 1.
Mechanisms of telomere extension and rDNA copy number maintenance that prevent cellular senescence and aging. (A) The budding yeast telomere structure consists of X′ and Y′ elements, and telomeric repeats. Core X that contains an autonomously replicating sequence (ARS) which is an origin of replication, and subtelomeric repeated elements (STR) that contains a Tbf1 binding site, are found in the X′ element. The ARS and Tbf1 binding site are also found in the Y′ element (Louis et al., 1994; Tham and Zakian, 2002; Wellinger and Zakian, 2012). (B) Telomerase-dependent pathway for telomere extension. Cdc13 and telomerase complex (Est1, Est2, Est3, and Tlc1) bind to both leading and lagging telomeres (Faure et al., 2010). DNA polymerase α (Polα)-primase complex generates RNA-DNA primers that initiate the synthesis of Okazaki fragments by DNA polymerase δ (Pol δ) (McElhinny et al., 2008; Perera et al., 2013) at lagging strand. After removal of the primers, the Okazaki fragments are ligated by DNA ligase I to form complementary lagging strand (Faure et al., 2010; Perera et al., 2013; Liu et al., 2017). Telomere extension mediated by telomerase occurs primarily at the leading telomere (Faure et al., 2010). The RNA-DNA primer is required for initiating synthesis of complementary leading strand by DNA polymerase ε (Pol ε) (Pursell et al., 2007; McElhinny et al., 2008; Perera et al., 2013). CST complex (Cdc13-Stn1-Ten1) bound at telomere end restricts the access of telomerase to telomere end. MRX complex (Mre11-Rad50-Xrs2) induces the binding of Tel1 (Nakada et al., 2003; McGee et al., 2010) to short telomere and executes 5′ to 3′ exonuclease activity to synthesize 3′ overhang (Diede and Gottschling, 2001). Both MRX and Tel1 promote Cdc13-mediated telomerase tethering to telomere (Tseng et al., 2006; Yang et al., 2017). Extension of telomere by telomerase is completed upon the synthesis of the CST complex (Pfeiffer and Lingner, 2013) and telomerase departure from the telomere. Polα-primase complex (Pol1, Pol12, Pri1, and Pri2) (Lue et al., 2014) interacts with CST complex at the telomere (Churikov et al., 2013) and generates an RNA-DNA primer for synthesis of complementary C-strand (Churikov et al., 2013; Pfeiffer and Lingner, 2013). (C) Homologous recombination pathways for telomere extension in the absence of telomerase or when telomerase is dysfunctional. The Rad51-dependent recombination pathway for the generation of type I survivors requires Exo1, Rad52 (Chen et al., 2001; Maringele and Lydall, 2004), Rad51, Rad54, and Rad57 (Chen et al., 2001; Claussin and Chang, 2015) while the Rad51-independent recombination pathway for the generation of type II survivors includes Exo1, Rad52 (Chen et al., 2001; Maringele and Lydall, 2004), Rad59, Sgs1, Def1, and MRX complex (Huang et al., 2001; Signon et al., 2001; Chen et al., 2005). (D) The structure of budding yeast rDNA (Poveda et al., 2010; Chand Dakal et al., 2016; Kobayashi and Sasaki, 2017). Abbreviations: ETS, external transcribed spacer; ITS, internal transcribed spacer; IGS, intergenic spacer; ARS, autonomous replication sequence; E-pro, rDNA non-coding promoter; RFB, replication fork barrier site; DSB, double-stranded DNA break. (E) Regulation of rDNA stability by homologous recombination (HR)-independent and -dependent pathways (unequal sister-chromatid or intrachromosomal recombination pathway) (Kobayashi and Sasaki, 2017; Sasaki and Kobayashi, 2017; Horigome et al., 2019; Morlot et al., 2019). Abbreviations: DSB, double-stranded break; ERC, extrachromosomal rDNA circles; SEP, senescence entry point; SSA, single strand annealing.