Abstract
Aims
We aimed to assess whether expression of whole‐blood RNA of sodium proton exchanger 1 (NHE1) and glucose transporter 1 (GLUT1) is associated with COVID‐19 infection and outcome in patients presenting to the emergency department with respiratory infections. Furthermore, we investigated NHE1 and GLUT1 expression in the myocardium of deceased COVID‐19 patients.
Methods and results
Whole‐blood quantitative assessment of NHE1 and GLUT1 RNA was performed using quantitative PCR in patients with respiratory infection upon first contact in the emergency department and subsequently stratified by SARS‐CoV‐2 infection status. Assessment of NHE1 and GLUT1 RNA using PCR was also performed in left ventricular myocardium of deceased COVID‐19 patients.
NHE1 expression is up‐regulated in whole blood of patients with COVID‐19 compared with other respiratory infections at first medical contact in the emergency department (control: 0.0021 ± 0.0002, COVID‐19: 0.0031 ± 0.0003, P = 0.01). The ratio of GLUT1 to NHE1 is significantly decreased in the blood of COVID‐19 patients who are subsequently intubated and/or die (severe disease) compared with patients with moderate disease (moderate disease: 0.497 ± 0.083 vs. severe disease: 0.294 ± 0.0336, P = 0.036). This ratio is even further decreased in the myocardium of patients who deceased from COVID‐19 in comparison with the myocardium of non‐infected donors.
Conclusions
NHE1 and GLUT1 may be critically involved in the disease progression of SARS‐CoV‐2 infection. We show here that SARS‐CoV‐2 infection critically disturbs ion channel expression in the heart. A decreased ratio of GLUT1/NHE1 could potentially serve as a biomarker for disease severity in patients with COVID‐19.
Keywords: COVID‐19, Infection, Heart, GLUT1, NHE1
Introduction
In the current SARS‐CoV‐2 pandemic, markers of increased risk for progression to severe disease are urgently needed to allocate limited healthcare resources to patients who need them most.
Furthermore, it is important to identify baseline medication that could be harmful (or beneficial) for patients with SARS‐CoV‐2 infection. The ion transporter sodium proton exchanger 1 (NHE1) is an important regulator of cellular pH in many tissues and is involved in the cellular response to stress, such as inflammation and infection. 1 , 2 Additionally, the glucose transporter 1 (GLUT1) is a critical glucose transporter in many tissues, especially the heart. 3
It has previously been shown that inhibitors of sodium–coglucose transporter 2 (SGLT2 inhibitors) may exert their beneficial effects directly on the heart via inhibition of NHE1, 4 , 5 as SGLT2 is not present in the heart. 6 Additionally, our working group has shown recently in that gliflozins increase myocardial expression of GLUT1, thus increasing glucose uptake into cardiomyocytes 7 while also altering myocardial sodium homoeostasis. 6 Furthermore, expression and activity of NHE1 have been shown to be affected by hypoxia and glucocorticoid levels, among others, via serum and glucocorticoid‐inducible kinase 1 (Sgk1). 8 , 9 It has been shown that GLUT1 activity (but not expression) is also stimulated by Sgk1. 10 Also, regulation of cellular pH is dependent on both GLUT1 expression and NHE1‐expression. 11 Interestingly, in the kidney, both low and high sodium levels can alter GLUT1 expression, 12 and NHE1‐deficient mice show strongly increased cardiac GLUT1 expression. 13 As such, it is reasonable that GLUT1 and NHE1 expression could be directly linked. We aimed to assess if SARS‐CoV‐2 infection altered the expression profile of these important cellular transporters in the blood of patients with COVID‐19. Furthermore, as COVID‐19 has been shown to affect cardiac function as well, we also investigated NHE1 and GLUT1 expression in the myocardium of deceased COVID‐19 patients.
We present data from our ongoing prospective clinical observational study investigating COVID‐19 patients at initial presentation to the emergency department (ED). We report on whole‐blood quantitative assessment of NHE1 and GLUT1 RNA using quantitative PCR (qPCR).
Methods
Patients were included in the study if they presented with signs of acute respiratory infection to the ED. Pre‐specified inclusion criteria were age ≥ 18 years and signs of acute respiratory infection, possibly COVID‐19. As all SARS‐CoV‐2‐positive patients reported symptomatic illness, the term COVID‐19 is used synonymously for these patients. In this study, we first included all patients with suspected COVID‐19 (defined by respiratory tract infection ± reported fever). Patients were then tested for SARS‐CoV‐2 infection. The patients with negative SARS‐CoV‐2 test result, who presented themselves with viral or bacterial respiratory tract infection, were used as control group.
Pre‐specified exclusion criteria were withdrawal of consent or patients unable to consent and delay of life‐saving diagnostic or therapeutic procedures by inclusion into the study (which did not occur in this study cohort). Researchers involved in the qPCR analysis were blinded with respect to group allocation/SARS‐CoV‐2 test status. As some patients were referred to our hospital ED from general practitioners with existing COVID‐19 diagnosis, nurses and physicians drawing blood and taking clinical data could not always be blinded to SARS‐CoV‐2 status, also for personnel safety considerations. However, all patients included into the study were treated equally with regard to diagnostic testing, timing of the testing, and safety precautions. The current study was approved by the Ethics Committee of the University of Regensburg. Informed consent was obtained from all patients.
After consent, clinical baseline characteristics and vital signs were documented for each patient. Furthermore, outcome and complications during hospital stay were assessed. Severe disease was defined as subsequent need for mechanical ventilation, admission to an intensive care unit, or death. Otherwise, patients were classified as moderate disease. An overview of inclusion procedures and patient stratification can be found in Figure 1 .
Figure 1.
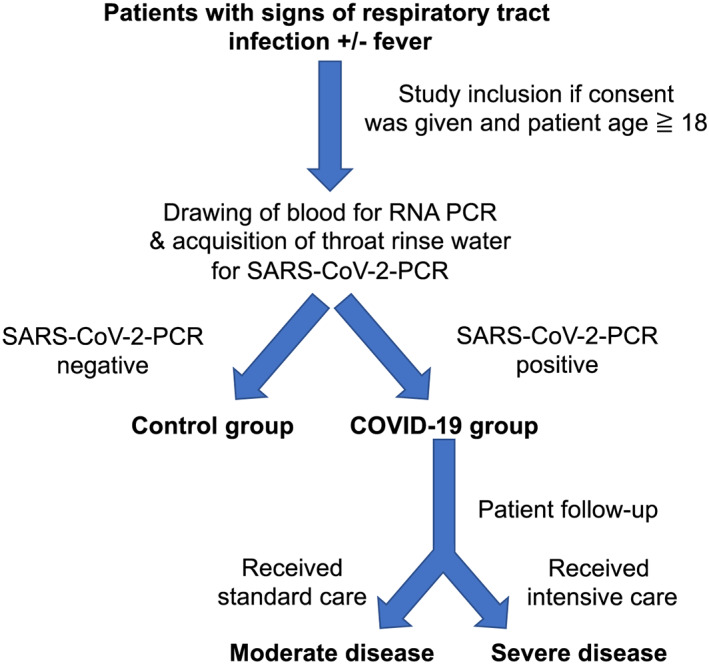
Overview of trial design. Patients were included in the study if they presented with signs of acute respiratory infection to the emergency department. Pre‐specified inclusion criteria were age ≥ 18 years and signs of acute respiratory infection. We included all patients with suspected COVID‐19 (defined by respiratory tract infection ± reported fever). For quantitative PCR analysis, patient blood was drawn by venepuncture from each consenting patient immediately after admission to the emergency department. Patients were then tested for SARS‐CoV‐2 infection. The patients with negative SARS‐CoV‐2 test result, who presented themselves with viral or bacterial respiratory tract infection, were used as control group. After consent, clinical baseline characteristics and vital signs were documented, and outcome and complications during hospital stay were assessed. Severe disease was defined as subsequent need for mechanical ventilation, admission to an intensive care unit, or death. Otherwise, (standard care) patients were classified as moderate disease.
For qPCR analysis, patient blood was drawn by venepuncture from each consenting patient immediately after admission to the ED and inactivated using Trifast (Ambion, Waltham, MA, USA). RNA was extracted using trichloromethane–chloroform solution and isopropanol solution. RNA was purified using the RNeasy Plus Mini Kit (Qiagen, Venlo, Netherlands) and transcribed to cDNA. Quantitative analysis of NHE1 and GLUT1 RNA was performed using the respective primer (NHE1: Hs00300047_m1, GLUT1: Hs00892681_m1, Applied Biosystems, Waltham, MA, USA) on a TaqMan apparatus (Applied Biosystems), and expression was normalized to β‐actin or GAPDH (β‐actin: Hs00357333_g1, GAPDH: Hs02786624_g1, Applied Biosystems). An extended description of the methodology can be found in the Supporting Information. We chose whole‐blood analysis for its easy applicability, requiring no separation stages of cells or plasma, and for safety considerations, as SARS‐CoV‐2 can easily be inactivated using Trifast without opening the blood tubes, thus avoiding potentially hazardous aerosol generation.
To strengthen our RNA data from whole blood of patients with COVID‐19, we also obtained left ventricular cardiac tissue from patients who died of COVID‐19 and analysed NHE1 and GLUT1 RNA in this cardiac tissue. Because of ethical and legal concerns, these deceased patients were not all from our ED study. However, infection had been verified by PCR from respiratory material in all of these patients, and all patients had symptomatic illness, which was determined to be the cause of death. As control tissue is extremely rare, we used a combination of left ventricular tissues from patients who had died from other respiratory infections or from patients whose hearts were destined for heart donations that ultimately could not be performed. No clinical data are available regarding the control group. Of note, there was no difference between the entities comprising the control group. As we had better experience with using GAPDH as a housekeeper for NHE1 in cardiac tissue, we used this instead of β‐actin for NHE1; however, to exclude this as confounder, we first analysed the ratio of GAPDH and β‐actin and found no significant difference (data not shown).
For SARS‐CoV‐2 testing, our facility mainly tested throat rinse water by PCR, but external test facilities also used throat swabs and sputum, which was accepted for our study if performed by a certified laboratory.
For statistical testing, normality was tested using the Kolmogorov–Smirnov test. For data with normal distribution, a Student's t‐test was performed in case of two groups without pairing. When testing multiple groups, an ANOVA was performed. For data for which normality could not be assumed, a Mann–Whitney U test was performed in case of two groups without pairing. Otherwise, a Kruskal–Wallis test was used. The respective post‐tests adjusting for multiple testing are referenced in the figure legends. Categorical data were tested using Fisher's exact test. The significance level was taken to 5% (two‐sided P). Data are presented as mean ± SEM, if not otherwise indicated. No data were excluded from the analysis.
Statistics were performed using GraphPad Prism v8 (GraphPad Software, San Diego, CA, USA) and SPSS 26. Original data can be made available in a blinded manner upon reasonable request.
Results
For this study, 43 patients were included in our analysis of whole‐blood RNA. In 21 of these patients (48.8%), SARS‐CoV‐2 infection could be confirmed. Clinical characteristics of these patients can be found in Table 1 .
Table 1.
Patient characteristics
| Control (n = 22) | SARS‐CoV‐2 (n = 21) | Statistics | |
|---|---|---|---|
| Baseline characteristics | |||
| Age | 57.1 ± 4.1 | 49.9 ± 3.6 | P = 0.2 a |
| Sex, % male (n) | 59.1 (13) | 57.1 (12) | P > 0.9 b |
| BMI | 26.9 ± 1.1 | 27.8 ± 1.2 | P = 0.6 a |
| Smokers (continued), % (n) | 9.1 (2) | 9.5 (2) | P > 0.9 b |
| Diabetes, % (n) | 27.3 (6) | 4.8 (1) | P = 0.09 b |
| Hypertension, % (n) | 50 (11) | 28.6 (6) | P = 0.2 b |
| Coronary artery disease, % (n) | 40.9 (9) | 4.8 (1) | P = 0.009 b |
| Chronic kidney disease, % (n) | 13.6 (3) | 9.5 (2) | P > 0.9 b |
| COPD, % (n) | 18.2 (4) | 0 (0) | P = 0.1 b |
| Asthma, % (n) | 13.6 (3) | 0 (0) | P = 0.2 b |
| Baseline medication | |||
| Baseline medication, % (n) | 68.2 (15) | 61.9 (13) | P = 0.8 b |
| ACE‐/AT1‐inhibitors, % (n) | 40.9 (9) | 19.1 (4) | P = 0.2 b |
| Beta‐blockers | 27.3 (6) | 19.1 (4) | P = 0.7 b |
| Aspirin (100 mg/day), % (n) | 36.4 (8) | 14.3 (3) | P = 0.2 b |
| Statins, % (n) | 40.9 (9) | 4.8 (1) | P = 0.009 b |
| Metformin, % (n) | 13.6 (3) | 0 (0) | P = 0.2 b |
| DPP4‐inhibitors, % (n) | 9.1 (2) | 0 (0) | P = 0.5 b |
| Sulfonylurea, % (n) | 0 (0) | 0 (0) | P > 0.9 b |
| Gliflozins, % (n) | 0 (0) | 0 (0) | P > 0.9 b |
| Incretins, % (n) | 0 (0) | 0 (0) | P > 0.9 b |
| Insulin, % (n) | 4.6 (1) | 4.8 (1) | P > 0.9 b |
| Immunosuppressants, % (n) | 13.6 (3) | 9.5 (2) | P > 0.9 b |
| Symptoms | |||
| Onset of symptoms to presentation (days), median (95% CI) | 3.5 (1; 8) | 8 (5; 13) | P = 0.01 c |
| Fatigue, % (n) | 90.9 (20) | 100 (22) | P = 0.5 b |
| Fever, % (n) | 72.7 (16) | 77.3 (17) | P > 0.9 b |
| Dyspnoea, % (n) | 63.6 (14) | 66.7 (14) | P > 0.9 b |
| Coughing, % (n) | 68.2 (15) | 71.4 (15) | P > 0.9 b |
| Chills, % (n) | 59.1 (13) | 61.9 (13) | P > 0.9 b |
| Physical markers | |||
| Body temperature (°C) | 37.6 ± 0.2 | 37.7 ± 0.2 | P = 0.8 c |
| SpO2 (%) | 95.3 ± 0.6 | 94.95 ± 0.8 | P = 0.7 a |
| Respiratory rate (/min) | 20.5 ± 1.3 | 22 ± 1.5 | P = 0.4 c |
| Heart rate (b.p.m.) | 89.2 ± 4.5 | 95.6 ± 3.3 | P = 0.3 a |
| Systolic blood pressure (mmHg) | 134.2 ± 5.2 | 132.9 ± 4.3 | P = 0.95 c |
| Diastolic blood pressure (mmHg) | 83.0 ± 2.9 | 80.4 ± 2.9 | P = 0.5 a |
| NEWS‐2 score | 3.1 ± 0.5 | 3.8 ± 0.6 | P = 0.4 c |
ACE, angiotensin‐converting enzyme; AT1, angiotensin II‐receptor‐subtype 1; BMI, body mass index; CI, confidence interval; COPD, chronic obstructive pulmonary disease; NEWS‐2, National Early Warning Score‐2.
t‐test.
Fisher's exact test.
Mann–Whitney U test.
Patients in the study were mostly male, and the mean age was 53.6 years, which was not significantly different between patients with confirmed SARS‐CoV‐2 infection and control. Body mass index was increased but similar in both groups. The median time from symptom onset to presentation in the ED differed significantly between the SARS‐CoV‐2 positive cohort and the control group. Clinical symptoms, vital signs, or National Early Warning Score‐2 (NEWS‐2) scoring on admission were not significantly different between both groups. In venous blood gas testing, pH, standard base excess, standard bicarbonate, and pCO2 did not differ between SARS‐CoV‐2 and control groups (Supporting Information, Table S1 ). In a non‐parametric correlation analysis, there was no significant association between NHE1 expression and pH in the SARS‐CoV‐2 or the control group.
In the SARS‐CoV‐2 group, one patient (4.8%), compared to six patients in the control group (27.3%), had diabetes mellitus, which was numerically different but did not reach statistical significance. Patients in the control group suffered significantly more often from coronary artery disease. Regarding drug therapy, only the usage of statins differed significantly between both groups.
NHE1 expression from whole blood was significantly elevated in patients with SARS‐CoV‐2 in comparison with control (P = 0.01; Figure 2 A ). There was no difference between male and female patients (P = not significant). RNA expression of GLUT1 was not significantly different between control group and COVID‐19 patients (Figure 2 B ).
Figure 2.
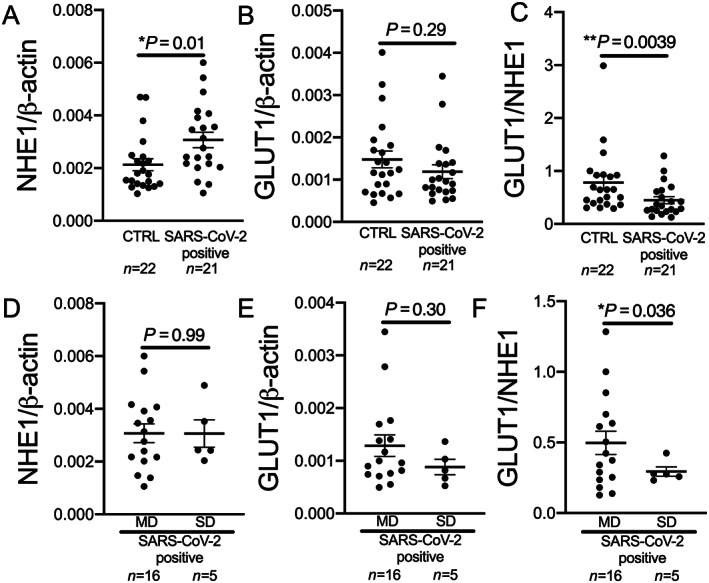
Sodium proton exchanger 1 (NHE1) and glucose transporter 1 (GLUT1) expression in the blood of COVID‐19 patients. (A) NHE1 expression is significantly increased in whole blood of patients with SARS‐CoV‐2 infection (SARS‐CoV‐2 positive) at first contact in the emergency department compared with control (CTRL; P = 0.01, Mann–Whitney U test). (B) GLUT1 expression is not significantly altered in whole blood of patients with SARS‐CoV‐2 infection compared with CTRL (P = 0.29, Mann–Whitney U test). (C) The ratio of GLUT1 and NHE1 is significantly decreased in patients with SARS‐CoV‐2 infection compared with CTRL (P = 0.0039, Mann–Whitney U test). (D and E) In patients with COVID‐19 at first contact in the emergency department, (D) NHE1 or (E) GLUT1 expression is not significantly different in patients who later develop moderate disease (MD) compared with patients later requiring intubation and/or dying [severe disease (SD); (D): P = 0.99, t‐test; (E): P = 0.30, t‐test]. (F) GLUT1 to NHE1 ratio is significantly decreased in patients with SD compared with patients with MD (P = 0.036, Welsh t‐test).
The ratio of GLUT1 and NHE1 whole‐blood RNA expression in SARS‐CoV‐2‐positive patients, however, was significantly reduced compared with control (Figure 2 C ).
In a stepwise multivariate linear regression analysis, only SARS‐CoV‐2 status was significantly associated with NHE1 expression (P = 0.015), in contrast to diabetes, gender, age, coronary artery disease, heart failure, obesity, arterial hypertension, hyperlipidaemia, smoking, and statin usage, which were non‐significant covariates (each P = not significant).
Five patients in the SARS‐CoV‐2 group had severe disease, and three of these five patients died. Neither NHE1 expression nor GLUT1 expression differed significantly between the COVID‐19 patients with moderate disease and the patients with severe disease (Figure 2 D,E ). The NEWS‐2 score was not different at first medical contact in the ED between COVID‐19 patients with moderate disease and patients with severe disease (moderate disease: 3.4 ± 0.7, severe disease: 4.8 ± 1.4, P = 0.26). The ratio of GLUT1 and NHE1 expression was significantly lower in patients with severe disease compared with moderate disease (Figure 2 F ).
RNA expression of NHE1 and GLUT1 was assessed in left ventricular myocardial tissue from patients who died of COVID‐19 (n = 6) and the control cohort (n = 7). Limited clinical data for the COVID‐19 patients are shown in Supporting Information, Table S2 . NHE1 expression was significantly elevated in patients who died from COVID‐19 compared with controls (Figure 3 A ). Furthermore, the expression of GLUT1 was significantly lower in the patients who had died from COVID‐19 compared with controls (Figure 3 B ). The ratio of GLUT1 to NHE1 was significantly lower in patients with COVID‐19 than in the control group (Figure 3 C ).
Figure 3.
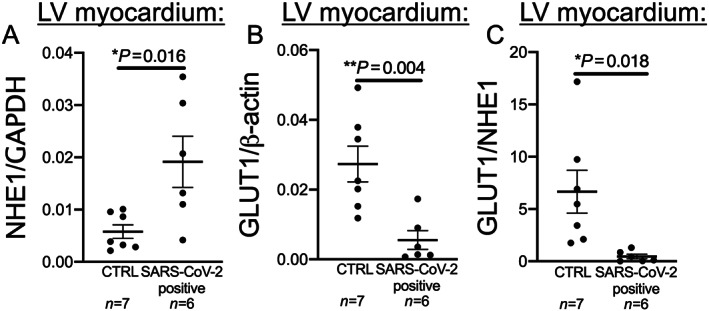
Sodium proton exchanger 1 (NHE1) and glucose transporter 1 (GLUT1) expression in left ventricular (LV) myocardium of deceased COVID‐19 patients. (A) In LV myocardium of deceased COVID‐19 patients (SARS‐CoV‐2 positive), NHE1 expression is significantly decreased compared with controls (CTRL; P = 0.016, t‐test). (B) In LV myocardium of deceased COVID‐19 patients (SARS‐CoV‐2 positive), GLUT1 expression is significantly decreased compared with CTRL (P = 0.004, t‐test). (C) Similar to the data from whole blood of patients in the emergency department, GLUT1/NHE1 ratio is significantly decreased in LV myocardium of deceased COVID‐19 patients (SARS‐CoV‐2 positive), compared with CTRL (P = 0.018, t‐test).
Discussion
For the first time, the current study shows that NHE1 RNA expression is up‐regulated in the blood of patients with COVID‐19 compared with other respiratory infections, which can be determined already at first contact in the ED. Furthermore, the ratio of GLUT1 to NHE1 is significantly lower in the blood of COVID‐19 patients who are subsequently intubated and/or die compared with moderately ill patients. Interestingly, this expression profile of NHE1 and GLUT1 extends beyond the blood, as expression of both transporters is significantly altered in left ventricular myocardium of deceased COVID‐19 patients. The ratio of GLUT1 and NHE1 in the myocardium of deceased COVID‐19 patients was significantly lower, resembling the lower ratio of GLUT1 and NHE1 in the blood of COVID‐19 patients in the ED, who are subsequently at risk of intubation or death. This suggests that NHE1 and GLUT1 may be critically involved in the disease progression of SARS‐CoV‐2 infection.
In COVID‐19, increased inflammatory markers such as C‐reactive protein and interleukin 6 have been reported, especially in severe COVID‐19 cases. 14 Cardiac complications, such as myocarditis, arrhythmias, acute coronary syndrome, or acute heart failure, are known complications in severe disease courses. Furthermore, elevated levels of cardiac markers, like cardiac troponin or brain natriuretic peptide, are common in these patients as surrogate of myocardial damage due to inflammation or ischaemia. 14 , 15 A potential explanation of the increased cardiovascular morbidity in COVID‐19 is the binding of SARS‐CoV‐2 to the host receptor angiotensin‐converting enzyme 2, which is highly expressed in the human heart and vascular cells, but exact pathophysiological processes of myocardial damage are still not fully understood.
NHE is an ion transporter protein family, which plays a critical role in regulation of intracellular pH or cell volume, and NHE1 is the predominant isoform of the NHE family in myocardial and vascular cells. 16 Previous studies found an increased activity of NHE1 associated with hypertrophy, cardiac fibrosis, and heart failure. 17 , 18 Therefore, NHE1 has been identified as a potential target in therapy of heart failure. 19 Recently, SGLT2 inhibitors, which decrease NHE1 activity, have been discussed as potential therapeutic agents in heart failure. 20 , 21 Interestingly, NHE1 activity seems to be involved in inflammatory response and leucocyte function. 22 , 23 In this study, we found elevated NHE1 expression in whole blood of patients with COVID‐19 in comparison with other bacterial or viral infections. This elevated expression was also present in myocardial tissue of deceased patients with COVID‐19. Of note, inflammatory parameters like interleukin 6, C‐reactive protein, or white blood cell count did not differ significantly between both groups (Supporting Information, Table S3 ). A potential confounder due to increased inflammatory response in the SARS‐CoV‐2 group cannot be excluded but seems therefore unlikely. Under the aspect of increased cardiovascular complications due to COVID‐19, the elevated expression of NHE1 could potentially be involved in pathophysiological pathways of cardiovascular damage due to COVID‐19. Increased NHE1 expression and/or activity could also be involved in or be dependent on altered platelet function in COVID‐19, 24 as NHE1 activation contributes to platelet activation. Interestingly, platelet activation can also be modulated by SGLT2 inhibitors. 25 In our COVID‐19 cohort, the platelet count is not significantly different between the control group and the COVID‐19 group (Supporting Information, Table S3 ). COVID‐19 patients with moderate disease and severe disease show no difference in platelet counts (moderate disease: 195.2 ± 14.84 vs. severe disease: 144.6 ± 35.25, t‐test P = 0.14, data expressed as mean ± SEM). Pearson correlations for platelet counts and NHE1 expression in the blood show neither a significant correlation in the control or COVID‐19 group nor when looking at COVID‐19 patients with moderate vs. severe disease. However, platelet count and platelet activity may of course differ, but as we used lysed whole blood for our experiments (safety for our personnel), we cannot determine platelet activity in our study cohort, as this would require fresh samples with high safety precautions. Nevertheless, after strong platelet activation, platelets aggregate and thus cannot be measured in the bloodstream; thus, platelet count is at least an indirect measure of platelet activity. 26
Interestingly, it has been shown that NHE1‐deficient mice show strongly increased cardiac GLUT1 expression. 13 GLUT1 is a glucose transporter of the GLUT family and ubiquitously expressed in almost all tissues. Primary function of GLUT1 is the basal cellular glucose uptake independent from insulin. Increased GLUT1 expression was shown in animal models of myocardial ischaemia or system inflammatory response syndrome, 27 , 28 suggesting a compensatory mechanism due to stress and hypoxia. While hypoxia can also increase NHE1 expression, among others, via Sgk1, 8 , 9 activity of the kinase has also been shown to alter GLUT1 activity. Further, GLUT1 expression was correlated with left ventricular contractility in that system inflammatory response syndrome animal model. Interestingly, a reduced myocardial GLUT1 expression was found in patients with end‐stage heart failure in one study. 29 In our study, GLUT1 whole‐blood RNA expression was not significantly different between COVID‐19 patients or control, but the ratio of GLUT1 to NHE1 was lower in patients with COVID‐19 in comparison with control. In patients with severe COVID‐19, the ratio was even lower than in patients with moderate disease. These altered expressions on NHE1 and GLUT1 were similar in myocardial tissue of deceased patients with COVID‐19. Furthermore, GLUT1 was significantly lower expressed in the myocardial tissue of patients who deceased due to COVID‐19. This pattern might reflect a maladaptation to inflammatory stress and myocardial ischaemia in patients with COVID‐19 and unfavourable disease progression. It may also hint at disturbed cellular ion handling and pH regulation, as both low and high sodium levels can alter GLUT1 expression, 12 and regulation of cellular pH is dependent on both GLUT1 expression and NHE1 expression and vice versa. 11
Further research regarding the role of these transporters is urgently warranted, especially regarding the potential effects of gliflozins in COVID‐19, which have been reported to affect both NHE1 and GLUT1. 4 , 7
Potential limitations of our study include the low number of diabetic patients in the COVID‐19 group, as diabetic patients are at increased risk of mortality when contracting SARS‐CoV‐2. Also, we could not investigate the effects of gliflozins on GLUT1 and NHE1 in COVID‐19 patients, which will need to be addressed in the future. Furthermore, an additional desirable control group for the analysis of cardiac expression of NHE1 and GLUT1 in deceased COVID‐19 patients would arguably be tissue from patients who have recovered from COVID‐19; however, this is of course not reasonably obtainable. In addition, these findings from a limited number of patients from only one site have to be validated in larger patient cohorts in the near future, also exploring whether reduced GLUT1/NHE1 ratio is an independent risk factor for mortality in COVID‐19.
Conflict of interest
The authors state that no conflict of interest exists.
Funding
J.M. is funded by the German Cardiac Society Clinician Scientist programme. L.S.M. is funded by the Deutsche Forschungsgemeinschaft (DFG) grant Ma1982/5‐1. S.W. and L.S.M. are funded by SFB 1350 TPA6 and the University of Regensburg ReForM C programme. S.W. is funded by DFG grants WA 2539/4‐1, 5‐1, 7‐1, and 8‐1.
Author contributions
J.M., J.H., and C.J. designed the study, gathered and analysed data, performed statistical analysis, wrote the manuscript, and are responsible for the integrity of the work as a whole. F.H., K.E., M.E., and C.M. gathered data and revised the manuscript for critical intellectual content. S.W., S.S., and L.S.M. analysed data and revised the manuscript for critical intellectual content.
Supporting information
Table S1. Venous blood gas analyses.
Table S2. Clinical data for deceased COVID‐19 patients with left ventricular tissue donation.
Table S3. Laboratory parameters.
Acknowledgements
We acknowledge the expert technical assistance of Teresa Stauber and Sebastian Benkhoff. Open access funding enabled and organized by Projekt DEAL.
Mustroph, J. , Hupf, J. , Hanses, F. , Evert, K. , Baier, M. J. , Evert, M. , Meindl, C. , Wagner, S. , Hubauer, U. , Pietrzyk, G. , Leininger, S. , Staudner, S. , Vogel, M. , Wallner, S. , Zimmermann, M. , Sossalla, S. , Maier, L. S. , and Jungbauer, C. (2021) Decreased GLUT1/NHE1 RNA expression in whole blood predicts disease severity in patients with COVID‐19. ESC Heart Failure, 8: 309–316. 10.1002/ehf2.13063.
[Correction added on 5 December 2020, after first online publication: Projekt Deal funding statement has been added.]
References
- 1. Deng X, Ji Z, Xu B, Guo L, Xu L, Qin T, Feng L, Ma Z, Fu Q, Qu R, Quo Q, Ma S. Suppressing the Na+/H+ exchanger 1: a new sight to treat depression. Cell Death Dis Nature Publishing Group UK 2019; 10: 370–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chai N, Bates P. Na+/H+ exchanger type 1 is a receptor for pathogenic subgroup J avian leukosis virus. Proc Natl Acad Sci 2006; 103: 5531–5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shao D, Tian R. Glucose transporters in cardiac metabolism and hypertrophy In Terjung R., ed. Comprehensive Physiology. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2015. p 331–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baartscheer A, Schumacher CA, Wüst RCI, Fiolet JWT, Stienen GJM, Coronel R, Zuurbier CJ. Empagliflozin decreases myocardial cytoplasmic Na+ through inhibition of the cardiac Na+/H+ exchanger in rats and rabbits. Diabetologia 2017; 60: 568–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Uthman L, Baartscheer A, Bleijlevens B, Schumacher CA, Fiolet JWT, Koeman A, Jancev M, Hollmann MW, Weber NC, Coronel R, Zuurbier CJ. Class effects of SGLT2 inhibitors in mouse cardiomyocytes and hearts: inhibition of Na+/H+ exchanger, lowering of cytosolic Na+ and vasodilation. Diabetologia 2018; 61( 3): 722–726. 10.1007/s00125-017-4509-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mustroph J, Wagemann O, Lücht CM, Trum M, Hammer KP, Sag CM, Lebek S, Tarnowski D, Reinders J, Perbellini F, Terracciano C, Schmid C, Schopka S, Hilker M, Zausig Y, Pabel S, Sossalla ST, Schweda F, Maier LS, Wagner S. Empagliflozin reduces Ca/calmodulin‐dependent kinase II activity in isolated ventricular cardiomyocytes. ESC Heart Fail 2018; 5: 642–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mustroph J, Lücht CM, Wagemann O, Sowa T, Hammer KP, Sag CM, Tarnowski D, Holzamer A, Pabel S, Beuthner BE, Sossalla S, Maier LS, Wagner S. Empagliflozin enhances human and murine cardiomyocyte glucose uptake by increased expression of GLUT1. Diabetologia 2019; 62: 726–729. [DOI] [PubMed] [Google Scholar]
- 8. Voelkl J, Pasham V, Ahmed MSE, Walker B, Szteyn K, Kuhl D, Metzler B, Alesutan I, Lang F. Sgk1‐dependent stimulation of cardiac Na+/H+ exchanger Nhe1 by dexamethasone. Cell Physiol Biochem 2013; 32: 25–38. [DOI] [PubMed] [Google Scholar]
- 9. Shimoda LA, Fallon M, Pisarcik S, Wang J, Semenza GL. HIF‐1 regulates hypoxic induction of NHE1 expression and alkalinization of intracellular pH in pulmonary arterial myocytes. Am J Physiol‐Lung Cell Mol Physiol Am Physiol Soc 2006; 291: L941–L949. [DOI] [PubMed] [Google Scholar]
- 10. Palmada M, Boehmer C, Akel A, Rajamanickam J, Jeyaraj S, Keller K, Lang F. SGK1 kinase upregulates GLUT1 activity and plasma membrane expression. Diabetes 2006; 55: 421–427. [DOI] [PubMed] [Google Scholar]
- 11. Lang KS, Mueller MM, Tanneur V, Wallisch S, Fedorenko O, Palmada M, Lang F, Bröer S, Heilig CW, Schleicher E, Weigert C. Regulation of cytosolic pH and lactic acid release in mesangial cells overexpressing GLUT1. Kidney Int 2003; 64: 1338–1347. [DOI] [PubMed] [Google Scholar]
- 12. Vestri S, Okamoto MM, de Freitas HS, Aparecida dos Santos R, Nunes MT, Morimatsu M, Heimann JC, Machado UF. Changes in sodium or glucose filtration rate modulate expression of glucose transporters in renal proximal tubular cells of rat. J Membr Biol 2001; 182: 105–112. [DOI] [PubMed] [Google Scholar]
- 13. Prasad V, Lorenz JN, Miller ML, Vairamani K, Nieman ML, Wang Y, Shull GE. Loss of NHE1 activity leads to reduced oxidative stress in heart and mitigates high‐fat diet‐induced myocardial stress. J Mol Cell Cardiol 2013; 65 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kermali M, Khalsa RK, Pillai K, Ismail Z, Harky A. The role of biomarkers in diagnosis of COVID‐19—a systematic review. Life Sci 2020; 254: 117788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. The European Society for Cardiology . ESC guidance for the diagnosis and management of CV disease during the COVID‐19 pandemic. 2020. https://www.escardio.org/Education/COVID‐19‐and‐Cardiology/ESC‐COVID‐19‐Guidance
- 16. Padan E, Landau M. Sodium‐proton (Na+/H+) antiporters: properties and roles in health and disease In Sigel A., Sigel H., Sigel R. K. O., eds. The Alkali Metal Ions: Their Role for Life. Cham: Springer International Publishing; 2016. p 391–458. [DOI] [PubMed] [Google Scholar]
- 17. Nakamura TY, Iwata Y, Arai Y, Komamura K, Wakabayashi S. Activation of Na+/H+ exchanger 1 is sufficient to generate Ca2+ signals that induce cardiac hypertrophy and heart failure. Circ Res 2008; 103: 891–899. [DOI] [PubMed] [Google Scholar]
- 18. Despa S, Bers DM. Na+ transport in the normal and failing heart—remember the balance. J Mol Cell Cardiol 2013; 61: 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baartscheer A, Schumacher CA, van Borren MMGJ, Belterman CNW, Coronel R, Opthof T, Fiolet JWT. Chronic inhibition of Na+/H+‐exchanger attenuates cardiac hypertrophy and prevents cellular remodeling in heart failure. Cardiovasc Res 2005; 65: 83–92. [DOI] [PubMed] [Google Scholar]
- 20. McMurray JJV, Solomon SD, Inzucchi SE, Køber L, Kosiborod MN, Martinez FA, Ponikowski P, Sabatine MS, Anand IS, Bělohlávek J, Böhm M, Chiang C‐E, Chopra VK, de Boer RA, Desai AS, Diez M, Drozdz J, Dukát A, Ge J, Howlett JG, Katova T, Kitakaze M, Ljungman CEA, Merkely B, Nicolau JC, O'Meara E, Petrie MC, Vinh PN, Schou M, Tereshchenko S, Verma S, Held C, DeMets D, Docherty KF, Jhund PS, Bengtsson O, Sjöstrand M, Langkilde AM, DAPA‐HF Trial Committees and Investigators . Dapagliflozin in patients with heart failure and reduced ejection fraction. N Engl J Med 2019; 381: 1995–2008. [DOI] [PubMed] [Google Scholar]
- 21. Zinman B, Wanner C, Lachin JM, Fitchett D, Bluhmki E, Hantel S, Mattheus M, Devins T, Johansen OE, Woerle HJ, Broedl UC, Inzucchi SE. Empagliflozin, cardiovascular outcomes, and mortality in type 2 diabetes. N Engl J Med 2015; 373: 2117–2128. [DOI] [PubMed] [Google Scholar]
- 22. Qadri SM, Su Y, Cayabyab FS, Liu L. Endothelial Na+/H+ exchanger NHE1 participates in redox‐sensitive leukocyte recruitment triggered by methylglyoxal. Cardiovasc Diabetol BioMed Central 2014; 13: 134–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao Y, Cui G, Zhang N, Liu Z, Sun W, Peng Q. Lipopolysaccharide induces endothelial cell apoptosis via activation of Na+/H+ exchanger 1 and calpain‐dependent degradation of Bcl‐2. Biochem Biophys Res Commun 2012; 427: 125–132. [DOI] [PubMed] [Google Scholar]
- 24. Manne BK, Denorme F, Middleton EA, Portier I, Rowley JW, Stubben CJ, Petrey AC, Tolley ND, Guo L, Cody MJ, Weyrich AS, Yost CC, Rondina MT, Campbell RA. Platelet gene expression and function in COVID‐19 patients. Blood 2020; 136: 1317–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Spigoni V, Fantuzzi F, Carubbi C, Pozzi G, Masselli E, Gobbi G, Solini A, Bonadonna RC, Dei Cas A. Sodium‐glucose cotransporter 2 inhibitors antagonize lipotoxicity in human myeloid angiogenic cells and ADP‐dependent activation in human platelets: potential relevance to prevention of cardiovascular events. Cardiovasc Diabetol BioMed Central 2020; 19: 46–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Würtz M, Hvas A‐M, Kristensen SD, Grove EL. Platelet aggregation is dependent on platelet count in patients with coronary artery disease. Thromb Res 2012; 129: 56–61. [DOI] [PubMed] [Google Scholar]
- 27. Rosenblatt‐Velin N, Montessuit C, Papageorgiou I, Terrand J, Lerch R. Postinfarction heart failure in rats is associated with upregulation of GLUT‐1 and downregulation of genes of fatty acid metabolism. Cardiovasc Res 2001; 52: 407–416. [DOI] [PubMed] [Google Scholar]
- 28. Bateman RM, Tokunaga C, Kareco T, Dorscheid DR, Walley KR. Myocardial hypoxia‐inducible HIF‐1α, VEGF, and GLUT1 gene expression is associated with microvascular and ICAM‐1 heterogeneity during endotoxemia. Am J Physiol‐Heart Circ Physiol Am Physiol Soc 2007; 293: H448–H456. [DOI] [PubMed] [Google Scholar]
- 29. Razeghi P, Young ME, Ying J, Depre C, Uray IP, Kolesar J, Shipley GL, Moravec CS, Davies PJA, Frazier OH, Taegtmeyer H. Downregulation of metabolic gene expression in failing human heart before and after mechanical unloading. Cardiology 2002; 97: 203–209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Venous blood gas analyses.
Table S2. Clinical data for deceased COVID‐19 patients with left ventricular tissue donation.
Table S3. Laboratory parameters.


