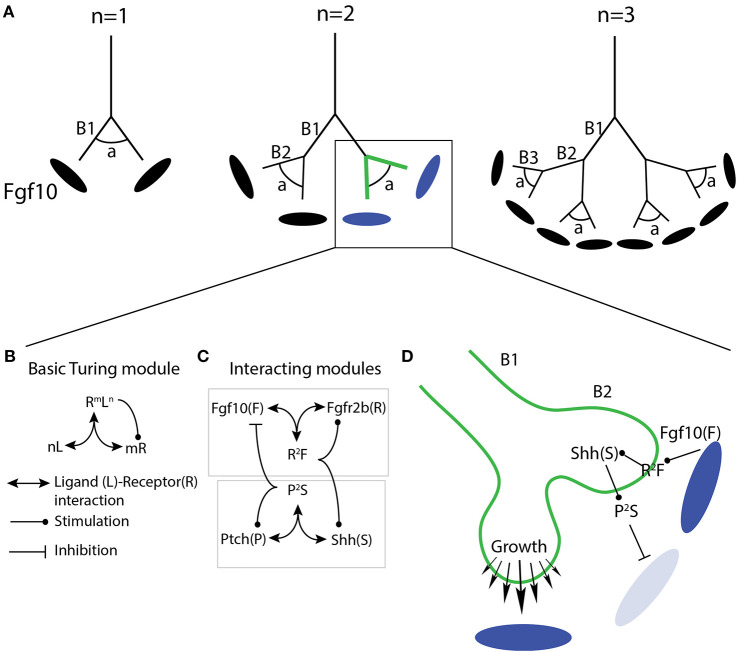
Figure 8.
Model of branching morphogenesis based on fractal geometry controlled by ligand-receptor based Turing mechanisms. (A) Three generations (n) of branch tip bifurcation following fractal rules wherein the angle of bifurcation (a) is consistent among generations, and the length of each successive branch (B) is shorter than the previous branch. Each tip grows toward a domain of Fgf10 expression. (B–D) A single branching event (box in ‘A’) is modeled using a ligand-receptor based Turing mechanism. (B) The basic Turing module contains n copies of a ligand (L) interacting with m copies of a receptor (R). The combined ligand-receptor interaction stimulates increased expression of the receptor. (C) Two Turing modules, Fgf10 and Shh, which interact in a negative feedback to control airway branching. Fgf10 combines with two copies of Fgfr2b, which stimulates expression of Fgfr2b and of Shh. Shh combines with two copies of Ptch, which stimulates expression of Ptch and represses expression of Fgf10. (D) Two daughter branches (B2) bifurcate from the parent branch (B1) as a result of the interacting ligand-receptor based Turing modules containing Fgf10 and Shh. Simulations using these two Turing modules predict the observed mesenchymal patterning of Fgf10 expression (blue pools), as well as predicting the observed growth fields (different sized arrows) of cultured embryonic lung explants. See text for further details.