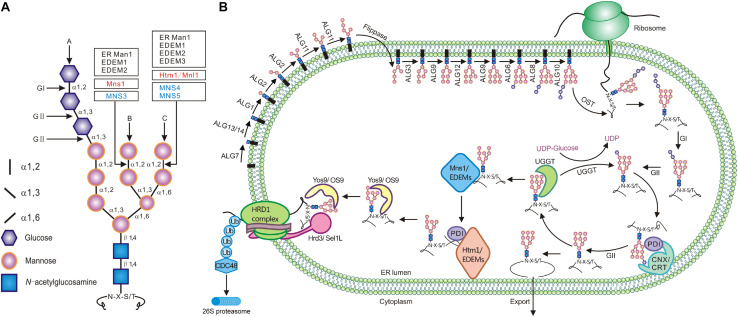
FIGURE 1.
The synthesis of N-glycan and its role in ERQC. (A) The structure of the three-branched Glc3Man9GlcNAc2. The inlet lists signs/shapes for different sugars and glycosidic linkages. Vertical arrows mark three different branches while the horizontal arrows indicate the cleavage sites of glucosidases and mannosidases. (B) The assembly of the N-glycan precursor starts at the cytosolic side of the ER membrane with addition of two GlcNAc residues to the membrane-anchored Dol-P linker followed by sequential attachment of the β1,4Man residue (catalyzed by ALG1), the α1,3Man/α1,6Man residues (by ALG2), and the two α1,2Man residues (by ALG11). The resulting Man5GlcNAc2-PP-Dol is flipped into the ER lumen where four additional Man residues are sequentially added from Dol-P-Man donor (catalyzed by ALG3, ALG9, ALG12, and ALG9) to generate Man9GlcNAc2-PP-Dol. ALG6, ALG8, and ALG10 catalyze the sequential addition of three Glc residues to form Glc3Man9GlcNAc2 that is en bloc transferred by OST onto certain Asn residues of nascent polypeptides. Immediately after the transfer, GI and GII rapidly remove the terminal and middle Glc residues to generate GlcMan9GlcNAc2. This N-glycan is recognized and bound by CNX/CRT that recruit additional chaperones and folding catalysts to assist the folding of those monoglucosylated glycoproteins. The removal of the last Glc residue by GII releases the glycoproteins from CNX/CRT. A correctly folded glycoprotein is demannosylated on the B-branch by Mns1/ERManI while trafficking to the Golgi apparatus whereas a misfolded/incompletely folded glycoprotein is recognized/bound by UGGT that adds back a Glc residue to regenerate GlcMan9GlcNAc2, forcing its reassociation with CNX/CRT for refolding. If a misfolded glycoprotein stays in the ER too long for engaging multiple futile refolding attempts, its N-glycans are slowly demannosylated by Mns1/ERManI and members of the Htm1/EDEM family (likely forming a disulfide bridged complex with members of the PDI family), generating N-glycans with an exposed α1,6Man residue. The ERAD lectin (OS-9/Yos9/EBS6) recognizes/binds α1,6Man-exposed N-glycans and works together with Hrd3/Sel1L/EBS5, which binds surface-exposed hydrophobic residues, to bring an irreparable misfolded glycoprotein onto the ER membrane-anchored ERAD complex containing a ubiquitin ligase (such as Hrd1) and its accessary factors. This complex not only ubiquitinates but also retrotranslocates a committed ERAD client that is subsequently escorted into the cytosolic proteasome for its complete degradation.