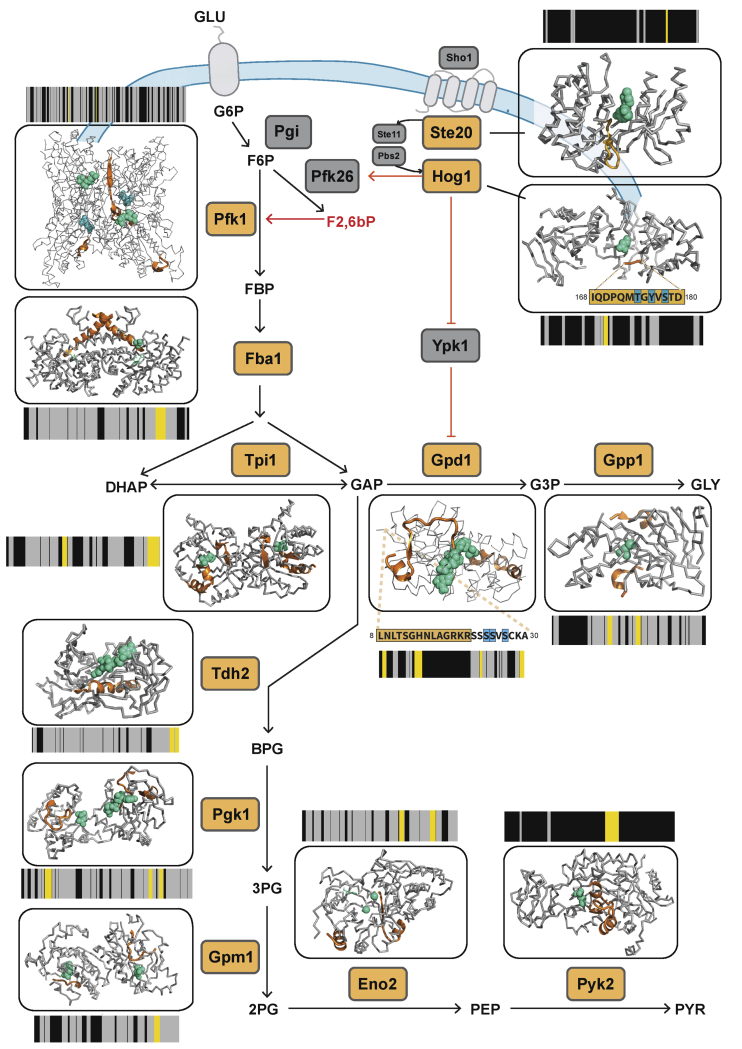
Figure 2.
Structural changes capture multiple regulatory events in yeast responding to osmotic shock
A schematic of the yeast HOG-MAPK pathway and its links to glycolytic and glycerol biosynthesis pathways. Proteins undergoing significant structural alterations upon osmotic shock are indicated with orange labels (|log2FC| >1, q-value < 0.05, two-sample t test with Storey method correction for multiple testing). The barcodes represent the changes in proteolytic fingerprints from N to C terminus. Each vertical bar represents a peptide that could be detected in samples subject to LiP. Peptides that changed in intensity between conditions are indicated by yellow (|log2FC| >1, q-value < 0.05), peptides detected by MS but unchanged between conditions are in gray, and peptides not detected by MS are in black. The structural models show changed LiP peptides (orange) mapped onto the 3D protein structures of yeast protein-metabolite complexes or evolutionary conserved holo-complex structures obtained by homology modeling; metabolites positioned in allosteric or active sites are indicated in green. For Hog1 and Gpd1, phosphorylation sites are indicated in blue on protein sequences. The allosteric regulator Fructose 2, 6-bisphosphate (F2,6bP), is depicted in red. The models shown are based on available structures: Pfk1 (PDB: 3o8o), Fba1 (PDB: 3qm3), Ste20 (PDB: 4zlo), Hog1 (PDB: 5ci6), Tpi1 (PDB: 1nf0), Gpd1 (PDB: 6e9o), Gpp1 (PDB: 2qlt), Tdh2 (PDB: 3pym), Pgk1 (PDB: 1qpg), Gpm (PDB: 1qhf), Eno2 (PDB: 1ebh), and Pyk2 (PDB: 1a3x).