Figure 3.
Molecular events underlying structural changes in the yeast proteome upon osmotic and heat shock
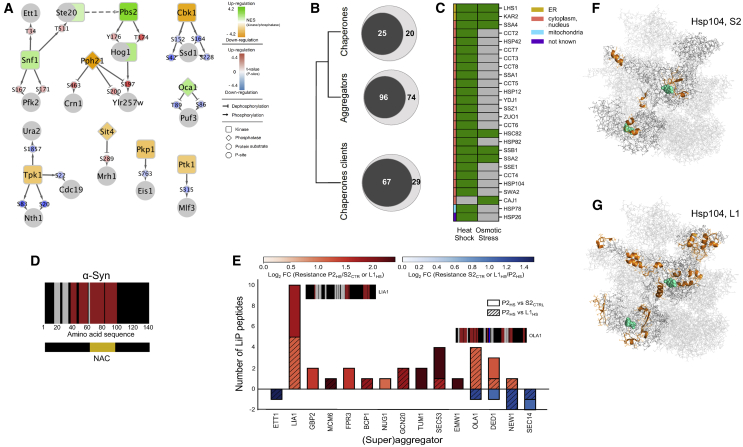
(A) Network representation of deregulated kinase activities and their target phosphosites on proteins showing structural changes upon osmotic shock of yeast cells. Structurally altered proteins are indicated by gray circles, kinases by squares, and phosphatases by diamonds; phosphorylation sites are indicated. Kinase and phosphatase activities are reported as normalized enrichment scores (NES), and phosphosite abundance changes are reported as p value-associated z-scores.
(B) Venn diagrams of the numbers of proteins of the indicated categories that are significantly structurally altered (|log2FC| >1, q-value < 0.05; two-sample t test with Storey method correction for multiple testing) after heat stress (inner circle) in relation with all detected proteins in that category (outer circle).
(C) Specific chaperones that show significant structural alterations (|log2FC| >1, q-value < 0.05; two-sample t test with Storey method correction for multiple testing) in heat or osmotic stress labeled by subcellular location.
(D) Structural barcode indicating differences in proteolytic resistance of alpha-synuclein fibrils versus monomer. Red/blue vertical bars indicate regions that show an increase/decrease in proteolytic resistance between fibril and monomer based on peptide intensity (|log2FC| >1, q-value < 0.05; Welch modified two-sample t test, p values adjusted for multiple testing with the Benjamini-Hochberg method). Detected peptides that do not change between conditions and non-detected peptides are plotted as grey and black bars, respectively. The aggregation core (NAC) is indicated.
(E) Bar plot showing protease resistance for all superaggregators and aggregators that become insoluble upon heat shock. The clear and hatched regions of the histograms show peptides indicative of increased/decreased (red/blue) proteolytic resistance for the indicated comparisons. The number of changed LiP peptides is plotted for each protein; hues indicate average strength of the fold change. Structural barcodes (as in D) are shown for selected proteins with large fold changes upon heat shock. Red/blue bars in the barcodes represent protein regions that increase/decrease proteolytic resistance in either of the two shown comparisons.
(F and G) LiP peptides (orange) of Hsp104 in (F) supernatant S2 and (G) whole-cell lysate L1 that change in response to heat shock mapped to the Hsp104 hexameric structure (PDB: 6n8t). ATP molecules binding to the chaperone catalytic site are depicted in cyan.