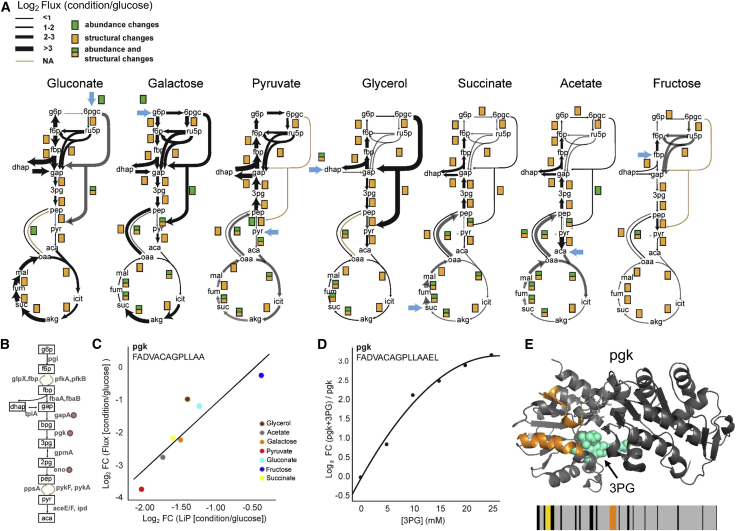
Figure 5.
Structural changes reflect functional flux alterations of E. coli metabolic enzymes
(A) 13C-based metabolic flux maps for E. coli grown in indicated nutrient conditions reported in Gerosa et al. (2015). The thickness of the black arrows indicates the flux fold change in relation to growth in glucose. Proteins with significant changes (|log2FC| >2, q-value < 0.05; p values adjusted for multiple testing with the Benjamini-Hochberg method) in abundance (green), structure (yellow), or both are indicated.
(B) Schematic of glycolytic enzymes. Red circles indicate enzymes with correlations between LiP peptide levels and metabolic flux.
(C) Linear regression between levels of the indicated LiP peptides derived from pgk and relative flux values through pgk across all nutrient conditions.
(D) Level of the best correlating LiP peptide of recombinant pgk spiked into an E. coli lysate with increasing 3-phosphoglycerate (3PG) concentration. This peptide is almost identical to the one correlated with flux across growth conditions in vivo (in [C]).
(E) The two LiP peptides that correlate with flux (orange) mapped onto the structure of pgk (PDB: 1zmr). 3PG bound to pgk is indicated in cyan. The barcode represents the change in proteolytic fingerprint along the sequence of pgk in galactose in relation to glucose (for barcodes corresponding to all growth conditions, see Figure S5C). Orange indicates peptides that change in intensity (|log2FC| >1, q-value < 0.05) between galactose and glucose, correlate with flux across all conditions, and correlate with substrate levels in an in vitro LiP experiment; yellow indicates peptides that change in intensity between galactose and glucose but do not meet the other two conditions; gray indicates peptides detected by MS that do not change between conditions; and black indicates peptides that are not detected by MS.
See also Table S3.