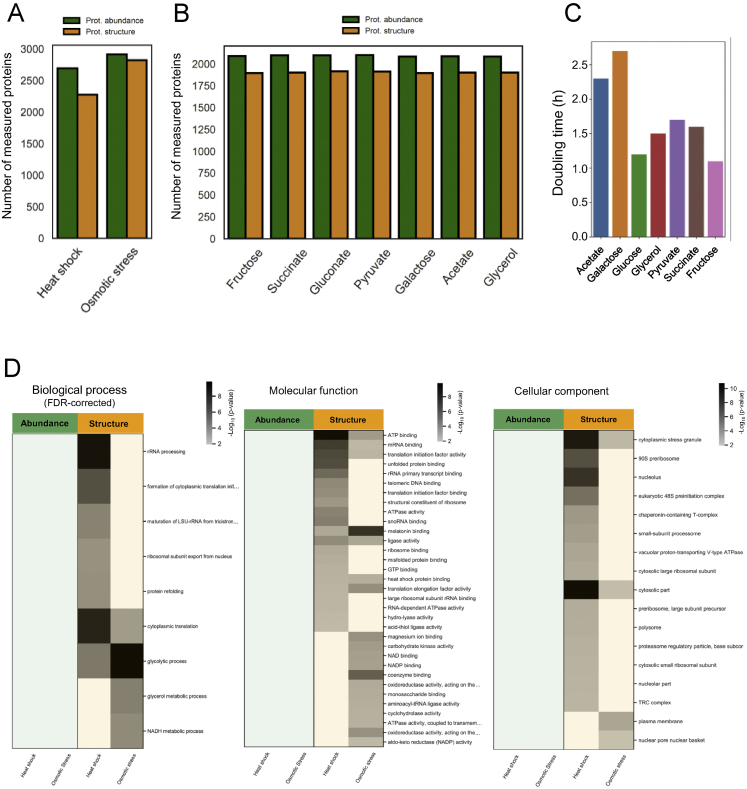
Figure S1.
Proteomic coverage and growth rate of bacteria in this study, and functional analysis of proteins that show structural and abundance changes during yeast response to acute stress, related to Figure 1
(A) The plot shows the number of yeast proteins detected by LC/MS-MS after digestion with trypsin only (which measures protein abundance, green bars) or upon limited proteolysis (which identifies structure-specific peptides, yellow bars), after the indicated stresses. (B) The plot shows the number of E. coli proteins detected by LC/MS-MS after digestion with trypsin only (which measures protein abundance, green bars) or upon limited proteolysis (which identifies structure-specific peptides, yellow bars) under the indicated conditions. (C) The plot shows the doubling time of E. coli in the seven different nutrient conditions used in this study. (D) The heat maps show functional categories (GO Biological Processes, Molecular Functions or Cellular Components) enriched among proteins that significantly change (|log2FC| >1, q-value < 0.05) in abundance (green) or structure (yellow) under the indicated stress conditions. P values for the enrichment (gray scale) were determined using Fisher’s exact test. Blank cells indicate molecular functions or cellular components that were not significantly enriched (i.e with p-value > 0.01) in a given condition. For the heat map showing Biological Processes, p-values were corrected for multiple hypothesis testing with the Benjamini-Hochberg method; blank cells indicate biological processes that were not significantly enriched (i.e with q-value > 0.05) in a given condition.