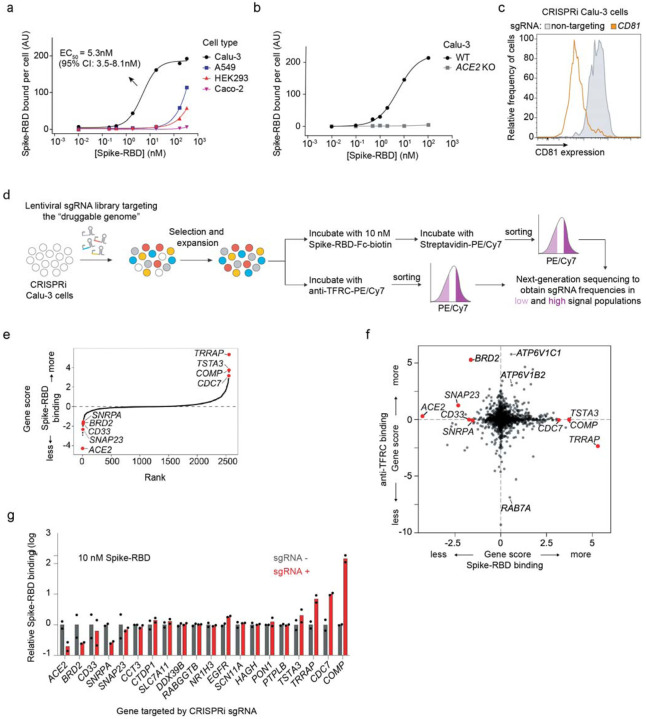
Figure 1: CRISPRi screen reveals cellular factors controlling Spike protein binding.
a, Human cell lines were incubated with different concentrations of recombinant SARS-CoV-2 Spike protein receptor-binding domain (Spike-RBD) and the amount of Spike-RBD bound per cell was quantified by flow cytometry. Only Calu3 cells (black) display saturable binding, for which an EC50 value was fit. Error of EC50 is the 95% confidence interval. b, Spike-RBD binding is eliminated in Calu-3 cells with ACE2 knockout (grey). c, Validation of CRISPRi activity of a Calu-3 CRISPRi line. Calu-3 cells stably expressing CRISPRi machinery were transduced with an sgRNA targeting CD81 (orange) or a non-targeting sgRNA (grey), and CD81 levels were determined by flow cytometry. d, CRISPRi screen strategy. CRISPRi Calu-3 cells transduced with an sgRNA library targeting the “druggable genome” are stained either with Spike-RBD or an anti-TFRC antibody. Cells are then FACS-sorted into bins based on Spike-RBD or anti-TFRC binding, and frequencies of cells expressing each sgRNA are determined for each bin by targeted next-generation sequencing. e, Rank-order plot of Spike-RBD hit genes. Genes selected for follow-up experiments are highlighted as red dots. f, Scatter plot of gene scores for the Spike-RBD screen (x-axis) and the anti-TFRC screen (y-axis). Genes selected for follow-up experiments are highlighted as red dots. g, Validation of hit genes using individually cloned sgRNAs. For each gene, the fold change of Spike-RBD binding at 10 nM Spike-RBD relative to cells not expressing sgRNAs is shown. Columns represent the average of two biological replicates (individual values shown as dots).