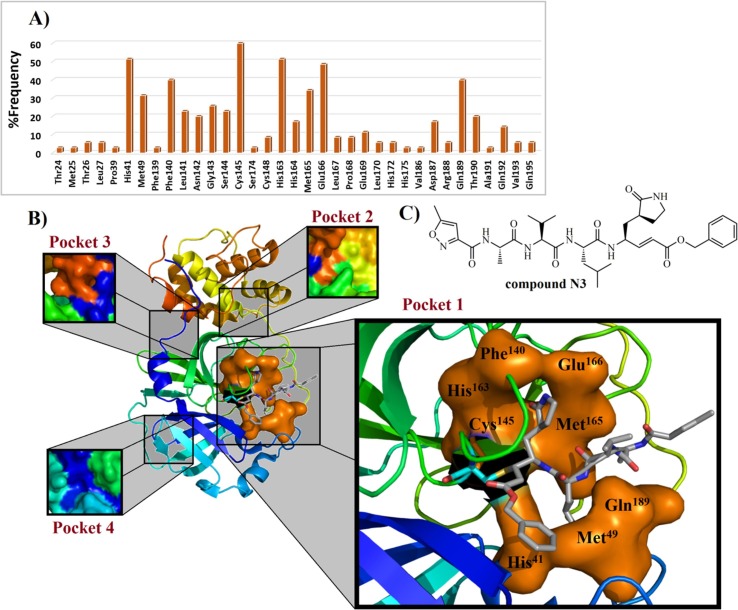
Fig. 14.
Frequency of residues present in ligand-3CLpro complexes (A) found in the literature and compound N3 in complex with eight most frequent amino acid residues of the pocket 1 (B) from SARS-CoV 3CLpro (PDB ID: 2HOB). In B), covalent Cys145 residue is shown as blue color, exhibiting its connectivity to Michael’s acceptor moiety. Finally, the most frequent amino acid residues from pocket 1 are demonstrated in a molecular surface overview (in orange color). In C), the Michael’s acceptor N3, a peptide-derived, which has been explored in different works, where it has been identified as a promising inhibitor of 3CLpro from MERS-CoV (IC50 = 0.28 µM),219 SARS-CoV-2 (EC50 = 16.77 µM),273 GS-WT12 (Ki = 9.0 ± 0.8 µM), WT-GPH6 (Ki = 2.3 ± 0.1 µM), and WT (Ki = 1.9 ± 0.1 µM) modified proteases.283.