Table 1.
Inhibitors targeting Coronaviruses found in the literature.
| Structure | Source | Organism | IC50 | Target | PDB - Interactions/(H-bond) | Ref. |
|---|---|---|---|---|---|---|
| Spike (S) protein | ||||||
| EEQAKTFLDKFNHEAEDLFYQSSGLGKGDFR | Synthetic | SARS-CoV | 0.1 µM | S protein | Not revealeda | 209 |
| Chymotrypsin-like cysteine protease (3CLpro) | ||||||
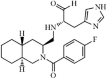 |
Synthetic | SARS-CoV | 57 µM | 3CLpro | (PDB: 4TWW) - Cys145, His41, Met49, Met165, Asp187, His163(H), Phe140, Leu141, and Glu166 | 210 |
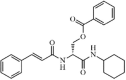 |
Synthetic | SARS-CoV | 30 µM | 3CLpro | (PDB: 3AW1) - Cys145(H) and Gln189(H) | 211 |
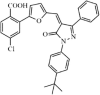 |
Synthetic | SARS-CoV | 5.8 µM | 3CLpro | (PDB: 2ALV) - Gly143(H), Ser144(H), Cys145(H), His163(H), His41(H), Leu27, Met49, and Gln189 | 212 |
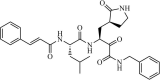 |
Synthetic | HCoV-NL63 | 1.08 µM | 3CLpro | (PDB: 6FV2) - His41(H), *Gly142(H), Phe139(H), and Glu166(H). | 213 |
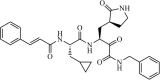 |
Synthetic | SARS-CoV | 0.24 µM | 3CLpro | (PDB: 5N5O) - Met49, Met165, Asp187, Gln189(H), and Thr190 | 213 |
 |
Synthetic | MERS-CoV | 1.7 µM | 3CLpro | (PDB: 4RSP) - Glu166(H), Glu169(H), Gln192(H), Cys148 (Covalent), and His41 | 214 |
| SARS-CoV | 0.2 µM | 3CLpro | Not revealeda | 214 | ||
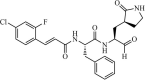
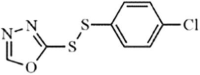 |
Synthetic | SARS-CoV | 0.51 µM | 3CLpro | (PDB: 2AMD) - Phe140, Leu141, His163, Met165, Glu166, His172, Asn142(H), Gly143(H), and Cys145(H) | 215 |
 |
Synthetic | MERS-CoV | 0.4 µM | 3CLpro | (PDB: 5WKL) - Cys148, Gln192(H), *Gln167(H), Glu169(H), His41(H), *His166(H), and *Phe143(H) | 216 |
| SARS-CoV | 5.1 µM | 3CLpro | Not revealeda | 216 | ||
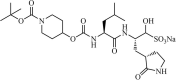
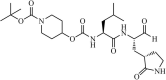 |
Synthetic | MERS-CoV | 0.6 µM | 3CLpro | Not revealeda | 216 |
| SARS-CoV | 2.1 µM | 3CLpro | Not revealeda | 216 | ||
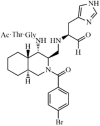 |
Synthetic | SARS-CoV | 26 µM | 3CLpro | (PDB: 4TWW) - Cys145, His163(H), Phe140, Leu141, and Glu166 | 217 |
 |
Synthetic | SARS-CoV | 95 µM | 3CLpro | Not revealeda | 218 |
 |
Synthetic | MERS-CoV | 0.28 µM | 3CLpro | (PDB: 1UK3) - Cys148 (Covalent), *His166(H), and His175(H) | 219, 220 |
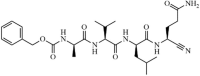 |
Synthetic | SARS-CoV | 4.6 µM | 3CLpro | (PDB: 3VB5) - Cys145(Covalent), Pro168, Thr190(H), Glu166(H), Gln189(H), His164(H), Phe140(H), and His163(H) | 221 |
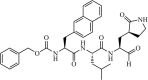 |
Synthetic | SARS-CoV | 0.23 µM | 3CLpro | Not revealeda | 222 |
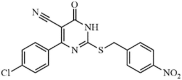 |
Synthetic | SARS-CoV | 6.1 µM | 3CLpro | (PDB: 1UK4) - Glu166(H), Gly143(H), Cys145(H), Met49, and Gln189 | 223 |
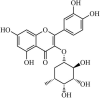 |
Synthetic | SARS-CoV | 24.1 µM | 3CLpro | Not revealeda | 224 |
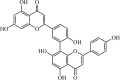 |
Natural | SARS-CoV | 8.3 µM | 3CLpro | (PDB: 2Z3E) - Leu141(H), His163(H), Val186(H), Gln189(H), and Gln192(H), | 225 |
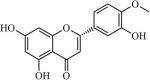 |
Natural | SARS-CoV | 8.3 µM | 3CLpro | Not revealeda | 226 |
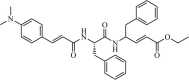 |
Synthetic | SARS-CoV | 1.0 µM | 3CLpro | Not revealeda | 227 |
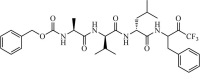 |
Synthetic | SARS-CoV | 10.0 µM | 3CLpro | (PDB: 1UK4) - His41, Met49, Phe140, Gly143(H), Cys145(H), His163, Met165, Glu166(H), Pro168, and Gln189(H) | 228 |
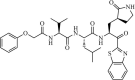 |
Synthetic | SARS-CoV | 1.7 µM | 3CLpro | Not revealeda | 229 |
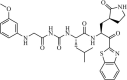 |
Synthetic | SARS-CoV | 10.0 µM | 3CLpro | (PDB: 1WOF) - Cys145, Ser144(H), His163(H), His164(H), Glu166(H), and Gln189(H) | 230 |
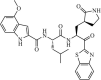 |
Synthetic | SARS-CoV | 0.74 µM | 3CLpro | (PDB: 1WOF) - Cys145, Ser144(H), His163(H), His164(H), Glu166(H), and Gln189(H) | 231 |
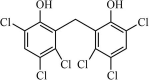 |
Synthetic | SARS-CoV | 5.0 µM | 3CLpro | (PDB: 1UK4) - Glu166(H), His163(H), Cys145(H), Ser144(H), Asn142(H), Phe140(H), Thr26(H), and His41 | 232 |
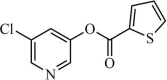 |
Synthetic | SARS-CoV | 0.5 µM | 3CLpro | Not revealeda | 233 |
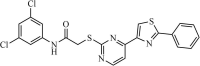 |
Synthetic | SARS-CoV | 3.0 µM | 3CLpro | Not revealeda | 234 |
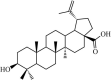 |
Natural | SARS-CoV | 10.0 µM | 3CLpro | Not revealeda | 235 |
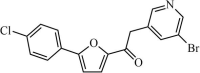 |
Synthetic | SARS-CoV | 13.0 µM | 3CLpro | Not revealeda | 236 |
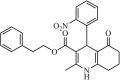 |
Synthetic | SARS-CoV | 8.9 µM | 3CLpro | Not revealeda | 237 |
| SARS-CoV | 2.5 µM | 3CLpro | Not revealeda | 238 | ||
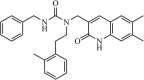 |
Synthetic | SARS-CoV | 17.2 µM | 3CLpro | Not revealeda | 239 |
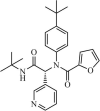 |
Synthetic | SARS-CoV | 1.5 µM | 3CLpro | Not revealeda | 240 |
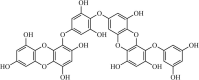 |
Natural | SARS-CoV | 2.7 µM | 3CLpro | (PDB: 2ZU5) - Thr190(H), His163(H), Ser144(H), His41(H), and Cys145(H) | 241 |
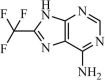 |
Synthetic | SARS-CoV | 13.9 µM | 3CLpro | Not revealeda | 242 |
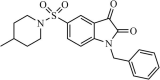 |
Synthetic | SARS-CoV | 1.04 µM | 3CLpro | (PDB: 1UK4) - Met49, Leu141, Asn142, Gly143(H), Ser144, Cys145(H), Met165, Arg188, Gln189, and Gln192 | 243 |
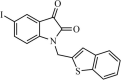 |
Synthetic | SARS-CoV | 0.95 µM | 3CLpro | (PDB: 1UK4) - Gly143(H), Cys145(H), His41(H), His164, Phe140, His163, and Met165 | 244 |
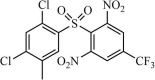 |
Synthetic | SARS-CoV | 0.3 µM | 3CLpro | (PDB: 1UK4) - Met49, His41(H), Pro39, Leu27, Cys145, His164, Met165, Leu167, Gln192, and Gln189 | 245 |
| Synthetic | SARS-CoV | 0.06 µM | 3CLpro | (PDB: 2A5A) - Glu189, Met49, His41, Cys145, Gly143(H), His163(H), Phe140, His172, and Glu166 | 246 | |
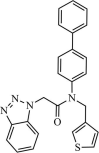 |
Synthetic | SARS-CoV | 0.051 µM | 3CLpro | Not revealeda | 247 |
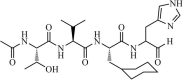 |
Synthetic | SARS-CoV | 0.098 µM | 3CLpro | (PDB: 3ATW) - Thr90(H), Glu166(H), His163(H), Phe140, Leu141, Asn142, His41, Met49, Met165, Asp187, and Cys145(H). | 248 |
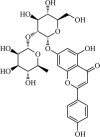 |
Natural | SARS-CoV | 27.5 µM | 3CLpro | (PDB: 4WY3) - Gln166(H), Leu167(H), Phe140(H), Gln189(H), Asn142(H), Thr26(H), and Thr24(H). | 249 |
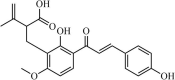 |
Natural | SARS-CoV | 11.4 µM | 3CLpro | (PDB: 2ZU5) - His163(H), Ser144(H), and Cys145(H) | 250 |
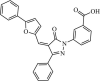 |
Synthetic | MERS-CoV | 5.8 µM | 3CLpro | (PDB: 4YLU) - His41, Ser147, Glu169(H), Leu170, Val193, and Gln195 | 212 |
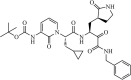 |
Synthetic | SARS-CoV2 | 0.67 µM | 3CLpro | (PDB: 6Y2F) - His41(H), Gly143(H), Cys145(H), Ser144(H), Phe140(H), Glu166(H), and His163(H) | 127 |
| SARS-CoV | 0.90 µM | 3CLpro | Not revealeda | 127 | ||
| MERS-CoV | 0.58 µM | 3CLpro | Not revealeda | 127 | ||
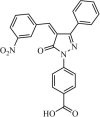 |
Synthetic | SARS-CoV | 9.6 µM | 3CLpro | (PDB: 1UK4) - Glu166(H), Gly143, Gln192, Met49, Arg188, and Gln189 | 251 |
| Papain-like protease (PLpro) | ||||||
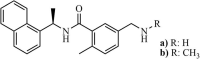 |
Synthetic | SARS-CoV | (a) 0.46 µM and (b) 1.3 µM | PLpro | (PDB: 3E9S) - Tyr269, Gln270, and Asp165 | 252 |
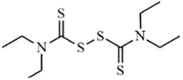 |
Synthetic | SARS-CoV | 14.2 µM | PLpro | (PDB: 4M0W) - Cys271 | 253 |
| MERS-CoV | 22.7 µM | PLpro | Not revealeda | 253 | ||
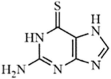 |
SARS-CoV | 5.0 µM | PLpro | Not revealeda | 254 | |
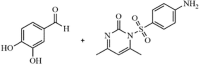 |
Synthetic | MERS-CoV | 6.6 µM | PLpro | (PDB: 4RF1) - Tyr279(H), Ser167(H), Pro163, Asp164, Asp165, *Gly248, *Thr249, Pro250, *Phe269, Glu273, Ala275, Val276, Gly277, and Thr308 | 255 |
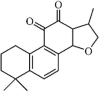 |
Natural | SARS-CoV | 0.8 µM | PLpro | Not revealeda | 256 |
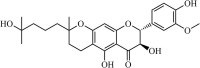 |
Natural | SARS-CoV | 5.0 µM | PLpro | Not revealeda | 257 |
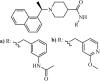 |
Synthetic | SARS-CoV | (a) 0.39 µM and (b) 0.35 µM | PLpro | (PDB: 3 MJ5) - Cys112, *Leu163, Asp165, Pro248, Pro249, Tyr265, Tyr269, Gln270, Tyr274, Thr302, and Asp303 | 258 |
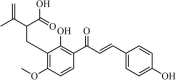 |
Natural | SARS-CoV | 1.2 µM | PLpro | (PDB: 3 MJ5) - His176(H) and His172(H) | 250 |
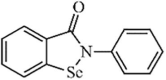 |
Synthetic | SARS-CoV | 8.45 µM | PLpro | (PDB: 2FE8) - His290, Asp287, His273, Ala289, Lys106(H), Cys112(Covalent), and Trp107(H). | 259b |
| SARS-CoV2 | 2.26 µM | PLpro | (PDB: 6W9C) - *Leu289, Ala288, Asp286, Lys105, Tyr268, *Trp106, His272, and *Cys111(Covalent). | 259b | ||
| NTPase/Helicase (nsP13) | ||||||
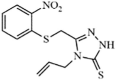 |
Synthetic | SARS-CoV | 6 µM | nsP13 | Not revealeda | 260 |
| 12 µM 50 µM 5.9 µM |
Y277AnsP13 K508AnsP13 WTnsP13 |
(homology model) Tyr277, Arg507, and Lys508. | 261 | |||
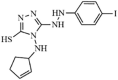 |
Synthetic | MERS-CoV | 2.5 µM | nsP13 | (homology model) Tyr7, Tyr159, Arg163, and Tyr171. | 261, 262 |
([Bi(L5) (H2O) (ClO4)3], L5 = 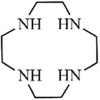
|
Synthetic | SARS-CoV | 5.0 µM | nsP13 | Not revealeda | 263 |
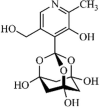 |
Synthetic | SARS-CoV | 3.0 µM | nsP13 | Not revealeda | 264 |
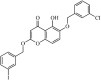 |
Synthetic | SARS-CoV | 11.0 µM | nsP13 | Not revealeda | 265 |
| Guanine-N7-Methyltransferase (also named as N7-MTase or nsP14) | ||||||
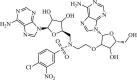 |
Synthetic | SARS-CoV | 0.6 µM | nsP14 | (PDB: 5C8T) - Trp385, Phe401, Tyr420, Phe426, Phe506, Cys387, Pro335, Val290, Trp292, Ile332, Phe367, Ala353, Val389, Asn386 (H), Arg310 (H), Gly333 (H), Ile338 (H), Lys336 (H), His424 (H), and Asp352 (H). | 266 |
*: Divergent residue labeling due to the difference between the SARS-CoV, SARS-CoV-2, and MERS-CoV proteins’ structures. a: The authors have not performed in silico studies. b: Preprint study published online at BioRxiv, doi: 10.1101/2020.05.17.100768, available at: https://www.biorxiv.org/content/10.1101/2020.05.17.100768v1.
