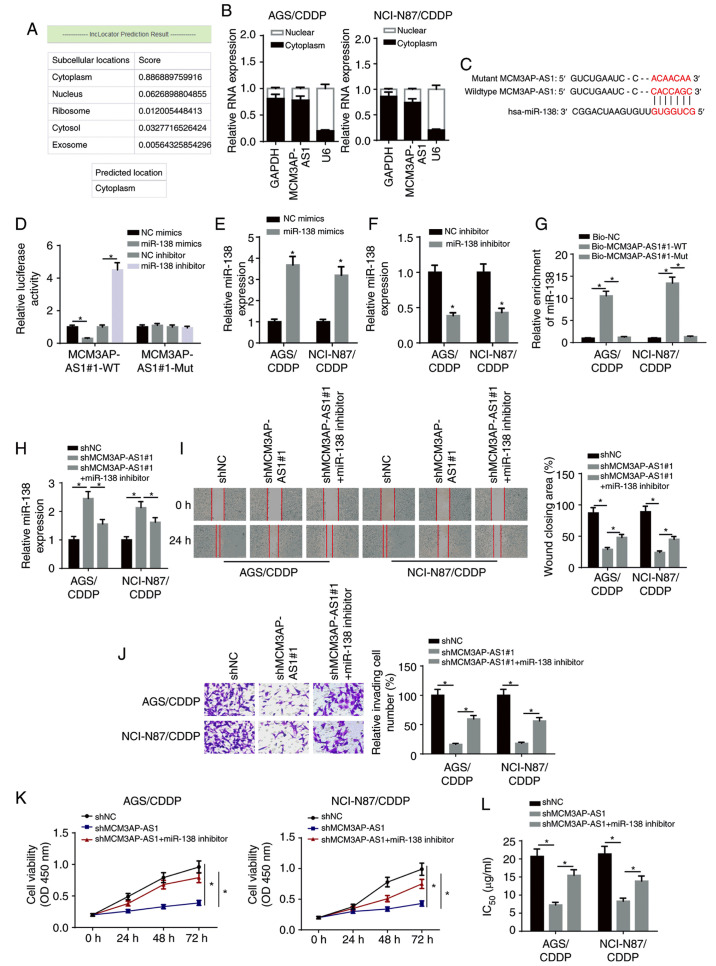
Figure 3.
MCM3AP-AS1 facilitates CDDP resistance of GC cells by targeting miR-138. (A) LncLocator program predicted the subcellular localization of MCM3AP-AS1. (B) MCM3AP-AS1 expression in the cytoplasmic and nuclear fractions was measured in NCI-N87/CDDP and AGS/CDDP cells by subcellular fraction assay. (C) The binding site between MCM3AP-AS1 and miR-138 was predicted using the starBase website. (D) The binding ability of MCM3AP-AS1 for miR-138 was detected in 293T cells by luciferase reporter assay. miR-138 expression in NCI-N87/CDDP and AGS/CDDP cells transfected with (E) NC mimics and miR-138 mimics, and (F) NC inhibitor and miR-138 inhibitor. (G) The binding capacity between MCM3AP-AS1 and miR-138 was verified by RNA pull-down assay. (H) miR-138 expression was assessed in shNC, shMCM3AP-AS1#1 and shMCM3AP-AS1#1+miR-138 inhibitor groups by reverse transcription-quantitative PCR. (I) Migratory (magnification, ×200) and (J) invasive abilities (magnification, ×200) of NCI-N87/CDDP and AGS/CDDP cells were detected in shNC, shMCM3AP-AS1#1 and shMCM3AP-AS1#1+miR-138 inhibitor groups by wound healing and Transwell assay, respectively. (K) Cell viability of NCI-N87/CDDP and AGS/CDDP cells was measured in shNC, shMCM3AP-AS1#1 and shMCM3AP-AS1#1+miR-138 inhibitor groups by Cell Counting Kit-8 assay. (L) The IC50 values of CDDP-resistant GC cells was tested in shNC, shMCM3AP-AS1#1 and shMCM3AP-AS1#1+miR-138 inhibitor groups by drug-sensitivity assay. *P<0.05. miR, microRNA; MCM3AP-AS1, MCM3AP antisense RNA 1; CDDP, cisplatin; GC, gastric cancer; sh, short hairpin RNA; NC, negative control; OD, optical density; WT, wild-type; Mut, mutant; Bio, biotinylated.