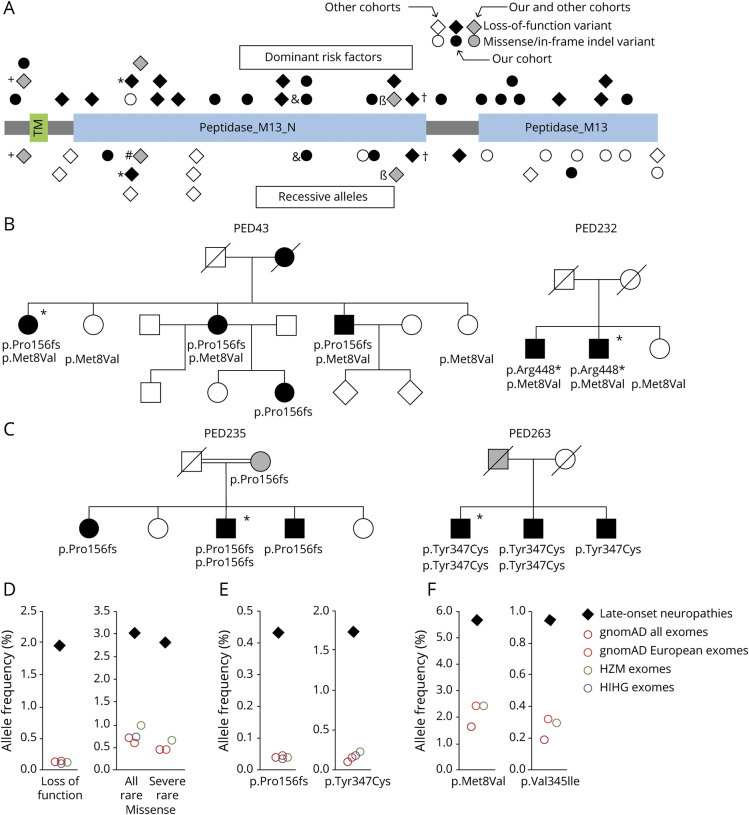
Figure 2. MME variants associated with late-onset neuropathies.
(A) Schematic representation of neprilysin and distribution of variants identified in this and other studies.10–12,25 Functionally relevant protein domains are indicated. Variants acting as autosomal-dominantly inherited risk factors are shown above and variants inherited as autosomal-recessive alleles are shown below the protein. Variants found in both groups of patients are shown twice and labeled with individual symbols (+, *, #, &, β, †). (B) Detection of serious MME variants and the p.Met8Val polymorphism in 2 pedigrees. Women are represented by circles; men are represented by squares. Symbols of affected individuals are filled (black, clinically affected; gray, probably affected by history or subclinical disease); those of unaffected individuals are empty. Crossed symbols represent deceased persons. Asterisks indicate index patients whose data are presented in tables S1, S3, and S4. (C) Pedigree of 2 families with both dominantly and recessively inherited late-onset CMT2 due to homozygous and heterozygous MME variant p.Pro156Leufs*14 and p.Tyr347Cys. (D) Comparison of cumulative allele frequencies for MME loss-of-function and rare missense variants between this study and whole-exome sequencing (WES) databases. Severe missense = predicted damaging/disease causing by at least 2 out of 3 in silico algorithms (PolyPhen-2, Scale-Invariant Feature Transform, Combined Annotation Dependent Depletion). (E) Individual frequencies of recurrent MME variants with a minor allele frequency (MAF) (gnomAD all exomes) between 0.0001 and 0.001 in this study and WES databases. (F) Individual frequencies of MME variants with an MAF (gnomAD all exomes) >0.001 in this study and WES databases. TM = transmembrane domain.