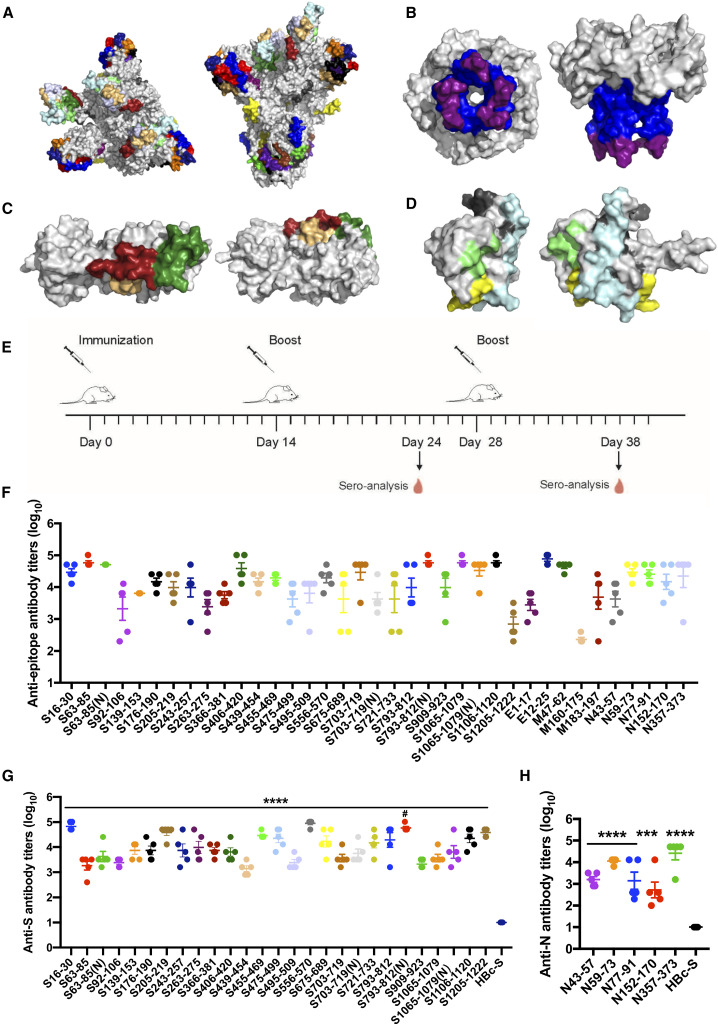
Figure 1.
Predication and validation of epitopes on SARS-CoV-2
(A–D) Molecular simulated structures and predicted epitopes of major proteins of SARS-CoV-2. Shown are top and side views of 3D structures (gray) and the predicted epitopes (colored) of the spike (S) protein (A), envelope (E) protein (B), membrane (M) protein (C), and nucleocapsid (N) protein (D).
(E–H) Epitope-conjugated HBc-S VLPs induce high antibody titers against epitope peptides and SARS-CoV-2 proteins.
(E) Schematic of the immunization design.
(F–H) 96-well plates were coated with peptides (F) and S (G) and N (H) proteins.
Data are shown as mean ± SEM (compared with the HBc-S control; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; one-way ANOVA followed by Dunnett’s test; compared with non-glycosylated epitope; #p < 0.05; Student’s t test).