Figure 1.
Mitochondrial ADP-Ribosylation Adapts in Response to Metabolic Changes
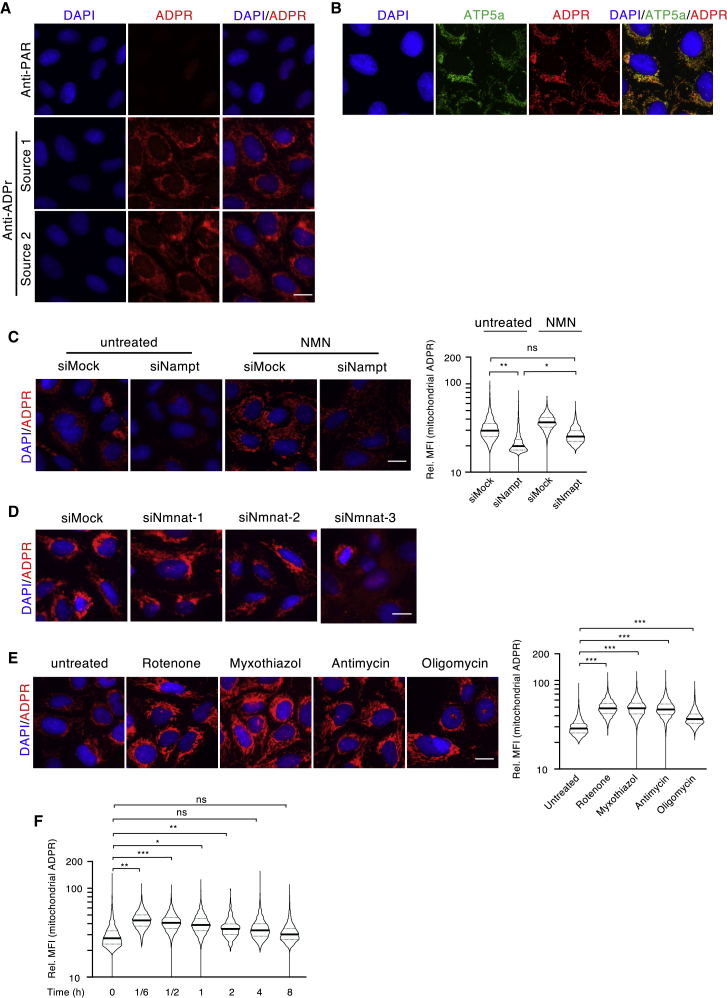
(A) Immunofluorescence (IF) staining of ADP-ribosylation (red) in U2OS cells using either an anti-PAR (upper panel) or two different pan-anti-ADPr antibodies (lower panels).
(B) Co-localization of ADP-ribosylation (red) and ATP5a (green).
(C) Influence of NAMPT knockdown and NMN supplementation on mitochondrial ADP-ribosylation. Left panel: representative IF images of the staining with an anti-ADPr antibody (red); right panel: quantification of the relative mean fluorescence intensity (Rel. MFI) of the mitochondrial ADP-ribosylation signal.
(D) Influence of NMNAT1, 2, or 3 on mitochondrial ADP-ribosylation.
(E) IF staining for ADP-ribosylation after treatment with inhibitors for the respiratory chain complexes and for the ATP synthase.
(F) Pulse-chase treatment of U2OS cells with rotenone for 10 min and recovery for the indicated time.
For representation of all quantifications, the signals for every event were normalized over the mean of the control/untreated, which was arbitrarily set to 30. The y axes of all violin plots are depicted as log10 scales. Scale bars indicate 20 μm. For statistical analysis, a Student’s t test was performed (n = 3–5; ∗p < 0.05; ∗∗p < 0.005; ∗∗∗p < 0.0005).