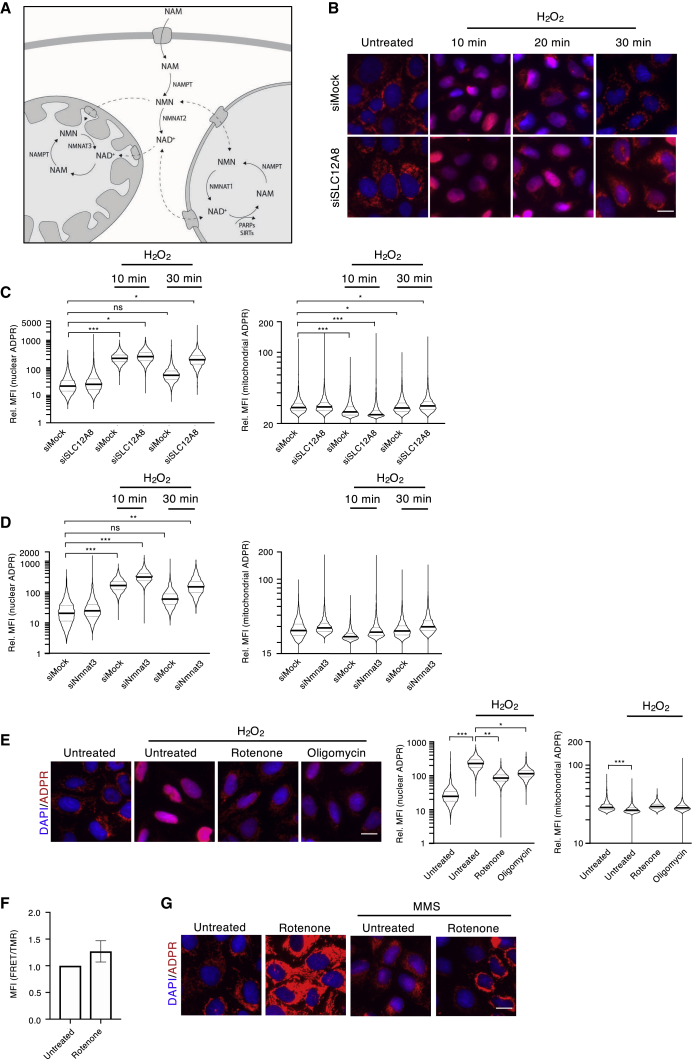
Figure 4.
Lack of NMNAT3 and the NMN Transporter SLC12A8 Extends Nuclear ADP-Ribosylation after H2O2 Treatment
(A) Schematic overview of compartmentalized NAD+ synthesis and breakdown in most transformed cells.
(B and C) U2OS cells were transfected with siRNA targeting SLC12A8, and nuclear (left) or mitochondrial (right) ADP-ribosylation was analyzed via IF and quantified at various time points following H2O2 treatment.
(D) NMNAT3 was knocked down in U2OS, and nuclear (left) or mitochondrial (right) ADP-ribosylation was analyzed via IF using an anti-ADPr antibody.
(E) U2OS cells were pre-treated for 1 h with rotenone or oligomycin prior to a 10-min treatment with H2O2 and nuclear and mitochondrial ADP-ribosylation was analyzed via IF.
(F) Mitochondrial NAD+ levels following rotenone treatment were assessed in stable U2OS Flp-In T-Rex cells expressing an inducible mitochondrial NAD+ sensor and analyzed via flow cytometry. Increased FRET ratios correspond to increased NAD+ levels. FRET ratios of three independent experiments including square deviation are shown.
(G) IF using the anti-ADPr antibody of U2OS cells pre-treated with rotenone and subsequently treated for 3 h with MMS. The quantifications of all signals (mitochondrial and nuclear) were normalized as described in Figure 1, and the y axes of all plots are depicted as log10 scale. Scale bars indicate 20 μm. For statistical analysis, a Student’s t test was performed (n = 3–5; ∗p < 0.05; ∗∗p < 0.005; ∗∗∗p < 0.0005).