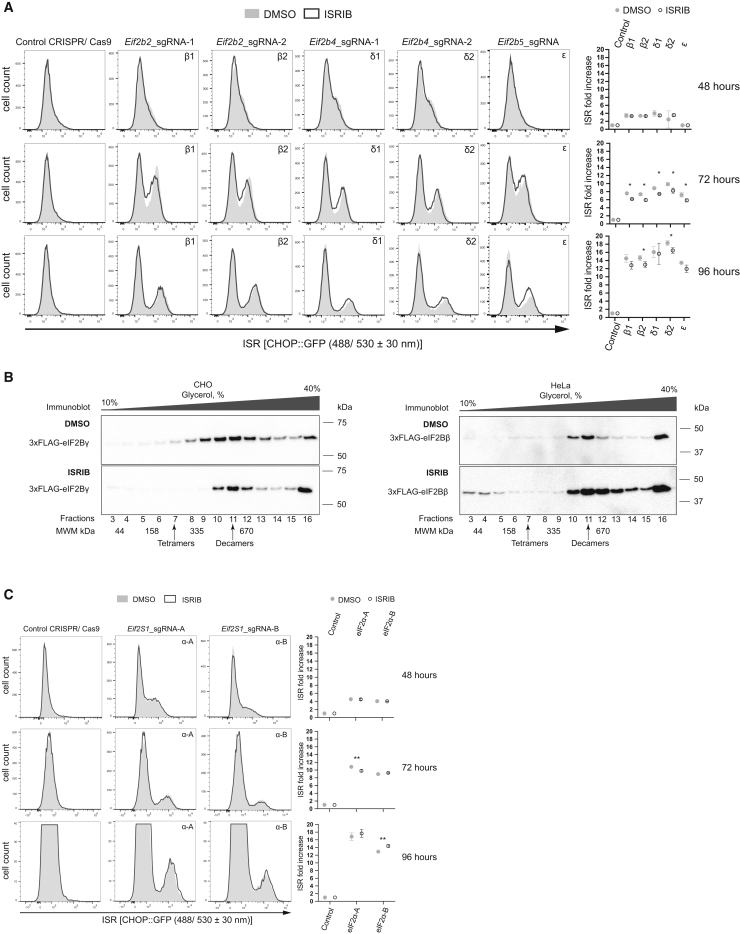
Figure 6.
Attenuated ISRIB Action in Cells Lacking eIF2(αP)
(A) Characterization of the ISR in Eif2S1S51A mutant CHO cells (lacking phosphorylatable eIF2α) depleted of eIF2B subunits by CRISPR/Cas9 targeting of their encoding genes. Two different guides for either beta (β1, β2) or delta (δ1, δ2) subunit, and one guide for epsilon subunit (ε) were transfected separately. Where indicated, the cells were exposed continuously to ISRIB (1 μM), commencing at the point of transduction with the CRISPR/Cas9 encoding plasmids and continued until harvest. Shown are histograms of the CHOP::GFP ISR reporter in populations of cells 48, 72, and 96 h following eIF2B gene targeting (±ISRIB) from a representative experiment performed three times. The mean ± SD (n = 3) of the ratio of fluorescent signal of the ISR-induced population (the right peak on the histograms) to the non-induced (left peak) are plotted to the right (∗p < 0.05 Student’s t test).
(B) Immunoblot of 3xFLAG-tagged endogenous eIF2Bγ detected with anti-FLAG M2 antibodies in CHO or 3xFLAG-tagged endogenous eIF2Bβ in HeLa cell lysates that were either treated with DMSO (top panel) or ISRIB (bottom panel) and resolved on a 10%–40% glycerol density gradient in a buffer of physiological salt concentration. The position of reference proteins of the indicated molecular weight in this gradient is indicated below the image and the arrows point to the predicted position of eIF2Bβδγε tetramers and eIF2B(α)2(βδγε)2 decamers.
(C) As in (A) above, but following CRISPR/Cas9-mediated depletion of eIF2B’s substrate eIF2, by targeting the Eif2S1 gene encoding its α-subunit. Two different guides, eIF2α-A and eIF2α-B, were transfected separately. Shown is a representative experiment performed twice. The mean ± SD (n = 2) of fluorescent signals ratio as in (A) are plotted to the right (∗∗p < 0.05 Student’s t test).