Abstract
Background
Viral load kinetics and duration of viral shedding are important determinants for disease transmission. We aimed to characterise viral load dynamics, duration of viral RNA shedding, and viable virus shedding of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in various body fluids, and to compare SARS-CoV-2, SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV) viral dynamics.
Methods
In this systematic review and meta-analysis, we searched databases, including MEDLINE, Embase, Europe PubMed Central, medRxiv, and bioRxiv, and the grey literature, for research articles published between Jan 1, 2003, and June 6, 2020. We included case series (with five or more participants), cohort studies, and randomised controlled trials that reported SARS-CoV-2, SARS-CoV, or MERS-CoV infection, and reported viral load kinetics, duration of viral shedding, or viable virus. Two authors independently extracted data from published studies, or contacted authors to request data, and assessed study quality and risk of bias using the Joanna Briggs Institute Critical Appraisal Checklist tools. We calculated the mean duration of viral shedding and 95% CIs for every study included and applied the random-effects model to estimate a pooled effect size. We used a weighted meta-regression with an unrestricted maximum likelihood model to assess the effect of potential moderators on the pooled effect size. This study is registered with PROSPERO, CRD42020181914.
Findings
79 studies (5340 individuals) on SARS-CoV-2, eight studies (1858 individuals) on SARS-CoV, and 11 studies (799 individuals) on MERS-CoV were included. Mean duration of SARS-CoV-2 RNA shedding was 17·0 days (95% CI 15·5–18·6; 43 studies, 3229 individuals) in upper respiratory tract, 14·6 days (9·3–20·0; seven studies, 260 individuals) in lower respiratory tract, 17·2 days (14·4–20·1; 13 studies, 586 individuals) in stool, and 16·6 days (3·6–29·7; two studies, 108 individuals) in serum samples. Maximum shedding duration was 83 days in the upper respiratory tract, 59 days in the lower respiratory tract, 126 days in stools, and 60 days in serum. Pooled mean SARS-CoV-2 shedding duration was positively associated with age (slope 0·304 [95% CI 0·115–0·493]; p=0·0016). No study detected live virus beyond day 9 of illness, despite persistently high viral loads, which were inferred from cycle threshold values. SARS-CoV-2 viral load in the upper respiratory tract appeared to peak in the first week of illness, whereas that of SARS-CoV peaked at days 10–14 and that of MERS-CoV peaked at days 7–10.
Interpretation
Although SARS-CoV-2 RNA shedding in respiratory and stool samples can be prolonged, duration of viable virus is relatively short-lived. SARS-CoV-2 titres in the upper respiratory tract peak in the first week of illness. Early case finding and isolation, and public education on the spectrum of illness and period of infectiousness are key to the effective containment of SARS-CoV-2.
Funding
None.
Introduction
Viral load kinetics and the duration of viral shedding are important determinants for disease transmission. They determine the duration of infectiousness, which is a critical parameter to inform effective control measures and disease modelling. Although several studies have evaluated severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) shedding, viral load dynamics and duration of viral shedding reported across studies so far have been heterogeneous.1 In several case series with serial respiratory sampling, peak viral load was observed just before, or at the time of, symptom onset.2, 3, 4 Viral RNA shedding was reported to be persistent in the upper respiratory tract and in faeces for more than 1 month after illness onset.1 However, the duration of SARS-CoV-2 RNA detection has not been well characterised. A comprehensive understanding of viral load dynamics, length of viral shedding, and how these measures relate to other factors, such as age and disease severity, is lacking.
We aimed to characterise the viral load dynamics of SARS-CoV-2, duration of viral RNA shedding by RT-PCR, and viable virus shedding in various body fluids, and to compare SARS-CoV-2 viral dynamics with those of SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV).
Research in context.
Evidence before this study
Understanding when patients are most infectious and the duration of infectiousness are of critical importance to controlling the COVID-19 pandemic. The duration of RNA detection across human coronaviruses has not been well characterised, and comprehensive understanding about viral load dynamics and the duration of viral shedding in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is lacking. We retrieved all articles reporting the dynamics and the duration of SARS-CoV-2, SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV) shedding in various specimens through systematic searches of major databases. Our research identified publications that included terms related to viral dynamics and viral shedding. We included case series, cohort studies, and randomised controlled trials in which the viral dynamics or the duration of viral shedding was reported. We excluded case reports, case series with fewer than five patients, and studies that did not have a clear time of symptom onset.
Added value of this study
To our knowledge, this is the first systematic review and meta-analysis that has examined and compared the viral dynamics of the three highly pathogenic human coronaviruses: SARS-CoV-2, SARS-CoV, and MERS-CoV. The results provide a comprehensive understanding regarding their viral kinetics and duration of shedding. Mean SARS-CoV-2 RNA shedding duration was 17·0 days (maximum shedding duration 83 days) in upper respiratory tract, 14·6 days (maximum 59 days) in lower respiratory tract, 17·2 days (maximum 35 days) in stool, and 16·6 days (maximum 60 days) in serum samples. Pooled mean SARS-CoV-2 shedding duration was positively associated with age. No study detected live virus beyond day 9 of illness, despite persistently high viral loads. SARS-CoV-2 viral load in the upper respiratory tract appeared to peak in the first week of illness, whereas SARS-CoV and MERS-CoV peaked later. Several studies reported similar viral loads at the start of infection among asymptomatic and symptomatic patients infected with SARS-CoV-2; however, most studies demonstrated faster viral clearance in asymptomatic individuals, as also seen in MERS-CoV, suggesting a shorter infectious period but with similar potential transmissibility at the onset of infection.
Implications of all the available evidence
Our study shows that despite evidence of prolonged SARS-CoV-2 RNA shedding in respiratory and stool samples, viable virus appears to be short-lived. Therefore, RNA detection cannot be used to infer infectiousness. High titres of SARS-CoV-2 are detected early in the disease course, with an early peak observed at the time of symptom onset to day 5 of illness; this finding probably explains the efficient spread of SARS-CoV-2 compared with SARS-CoV and MERS-CoV. This has important implications for SARS-CoV-2 transmission in the community and hospital setting, emphasising the importance of early case finding and prompt isolation as well as public education about the spectrum of illness. Our study shows that isolation practices should be commenced with the start of first symptoms, which can include mild and atypical symptoms, preceding typical symptoms of COVID-19 such as cough and fever. However, given the potential delays in isolation of patients, even the early detection and isolation strategy might not be fully effective in containing SARS-CoV-2.
Methods
Search strategy and selection criteria
We retrieved all English-language research articles reporting viral dynamics or the duration of shedding of SARS-CoV-2, SARS-CoV, or MERS-CoV in various specimens through systematic searches of major databases, including MEDLINE, Embase, Europe PubMed Central, medRxiv, and bioRxiv, and the grey literature from Jan 1, 2003, to June 6, 2020, using medical subject headings terms (appendix p 14). We also manually screened the references of included original studies to obtain additional studies. Studies published before 2003 were excluded because the first recognised case of SARS-CoV was identified in March, 2003.
Studies were eligible if they met the following inclusion criteria: report on SARS-CoV-2, SARS-CoV, or MERS-CoV infection, and report viral load kinetics, duration of viral shedding, or viable virus shedding. We excluded review papers; animal studies; studies on environmental sampling; case reports and case series with less than five participants, due to likely reporting bias; papers in which the starting point of viral shedding was not clear or reported from post hospital discharge; and modelling studies with no original data.
Data extraction
Two authors (MT and OL) screened and retrieved articles according to the eligibility criteria. Four reviewers (MT, OL, JS, and MC) reviewed full-text articles and selected articles to be included. From each study, the following variables were extracted as a minimum: name of first author, year of publication, city and country, sample size, median age, sex ratio, time from symptom onset to viral clearance detected by RT-PCR and culture in different specimens, and longest reported time to viral clearance. If these data were not reported, we also contacted the authors to request the data. If available, we extracted data on peak viral load, clinical outcome, and reported factors associated with duration of viral shedding.
Two authors (OL and JS) independently assessed study quality and risk of bias using the Joanna Briggs Institute Critical Appraisal Checklist tools,5 which comprise standardised checklists, for the different study designs included in this review. Any disagreements regarding grading of quality were resolved through discussion with a third author (MC).
Data analysis
For every study included, we calculated the mean duration of viral shedding and 95% CIs. We applied the random-effects model to estimate a pooled effect size. We generated forest plots to show the detailed representation of all studies based on the effect size and 95% CI. If not reported, we derived means and SDs from sample size, and median, IQR, minimum, and maximum values.6 Heterogeneity between studies was quantified by the I 2 index and Cochran's Q test. We did not assess publication bias because usual appraisal methods are uninformative when meta-analysed studies do not include a test of significance. We used a weighted meta-regression with an unrestricted maximum likelihood model to assess the effect of potential moderators on the pooled effect size (p<0·05 was considered to be significant). The eligibility criterion for meta-regression was the presence of at least ten studies (referring to one virus) for each covariate. All statistical analyses were done with Comprehensive Meta-Analysis (version 3) software (Biostat, Englewood, NJ, USA).
This systematic review is registered with PROSPERO, CRD42020181914, and will be updated periodically.
Role of the funding source
There was no funding source for this study. The corresponding author and senior author (AH) had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
The systematic search identified 1486 potentially relevant articles. 350 articles were retrieved for full-text review. After reviewing the eligibility criteria, 79 studies (5340 individuals) on SARS-CoV-2,2, 3, 4, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82 eight (1858 individuals) on SARS-CoV,83, 84, 85, 86, 87, 88, 89, 90 and 11 (799 individuals) on MERS-CoV91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101 were included (figure 1 ).
Figure 1.
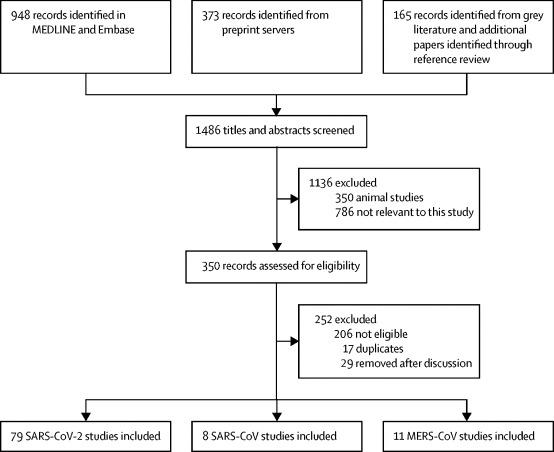
Study selection
MERS-CoV=Middle East respiratory syndrome coronavirus. SARS-CoV=severe acute respiratory syndrome coronavirus. SARS-CoV-2=severe acute respiratory syndrome coronavirus 2.
Of the 79 papers included, 58 studies were done in China (appendix p pp 1–4).2, 10, 11, 12, 14, 15, 16, 17, 19, 20, 21, 23, 24, 25, 26, 27, 28, 35, 36, 37, 38, 39, 40, 41, 43, 44, 48, 49, 50, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 65, 66, 67, 68, 69, 70, 71, 72, 73, 75, 76, 77, 78, 79, 80, 81, 82 73 studies included only patients who were admitted to hospital.3, 4, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 31, 32, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 52, 53, 54, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82 Six studies reported viral load dynamics exclusively in children (age younger than 16 years).7, 8, 9, 10, 11, 12 Two additional studies included children, but data on viral load dynamics were presented in aggregate with adults.13, 14
61 studies reported median or maximum viral RNA shedding in at least one body fluid and were eligible for quantitative analysis,3, 4, 7, 9, 10, 11, 12, 14, 15, 16, 17, 19, 20, 21, 22, 24, 25, 27, 28, 29, 30, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 45, 47, 48, 50, 51, 53, 54, 57, 58, 59, 60, 61, 62, 63, 64, 65, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 78, 79, 80, 82 and six studies provided duration of shedding stratified by illness severity only.13, 39, 52, 55, 77, 81 Of those studies, 43 (including 3229 individuals) reported duration of shedding in the upper respiratory tract (mean viral shedding duration 17·0 days [95% CI 15·5–18·6]; figure 2 ), seven (260 individuals) in the lower respiratory tract (14·6 days [9·3–20·0]; appendix p 6), 13 (586 individuals) in stool samples (17·2 days [14·4–20·1]; appendix p 7), and two (108 individuals) in serum samples (16·6 days [3·6–29·7]; appendix p 6). Maximum duration of RNA shedding reported was 83 days in the upper respiratory tract,35 59 days in the lower respiratory tract,27 126 days in stool samples,88 and 60 days in serum samples.78
Figure 2.
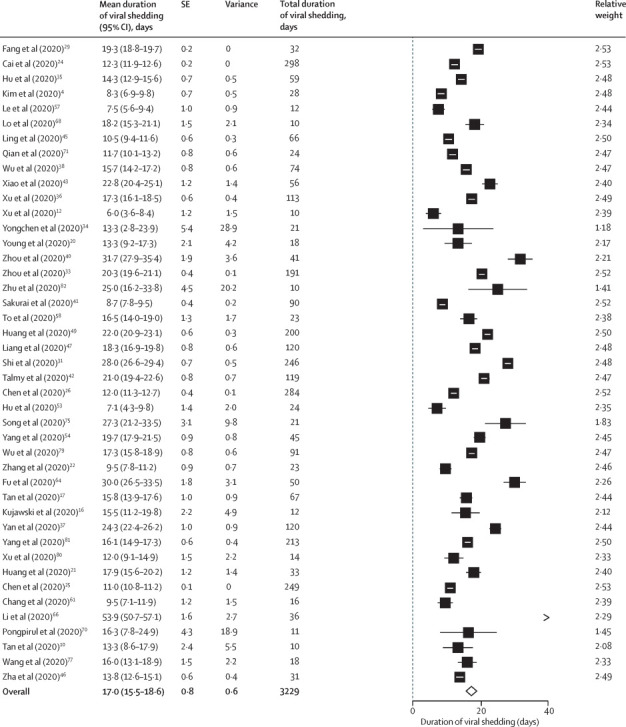
Pooled mean duration (days) of SARS-CoV-2 shedding from the upper respiratory tract (random-effects model)
SARS-CoV-2=severe acute respiratory syndrome coronavirus 2.
Studies reporting duration of viral shedding in upper respiratory tract and stool samples were eligible for meta-regression analysis. Pooled mean viral shedding duration was positively associated with age (slope 0·304 [95% CI 0·115–0·493]; p=0·0016), but not sex (p=0·28; appendix pp 7–8). When adjusted for the proportion of males in a multivariable analysis, mean age was positively associated with the mean duration of viral shedding in upper respiratory tract specimens (p=0·0029). There was a positive but non-significant association between mean age and duration of shedding in stool samples (p=0·37; appendix p 8).
Eight of 13 studies evaluating SARS-CoV-2 viral load in serial upper respiratory tract samples showed peak viral loads inferred from cycle threshold values within the first week of symptom onset.2, 3, 4, 8, 15, 16, 17, 18, 19, 20, 21, 22, 23 The highest viral loads were reported soon after or at the time of symptom onset,2, 8, 15, 21, 23 or at day 3–5 of illness,3, 4, 20 followed by a consistent decline.
Five studies that evaluated viral load dynamics in lower respiratory tract samples observed a peak viral load in the second week of illness.3, 4, 17, 21, 23 By contrast, the dynamics of SARS-CoV-2 shedding in stool samples was erratic, with highest viral loads reported on day 7,17 2–3 weeks,22, 23 and up to 5–6 weeks after symptom onset.21 Although two studies reported significantly higher viral titres in stool samples than in respiratory samples,8, 23 one study reported lower viral load in stool samples than in both lower and upper respiratory tract samples at the time of symptom onset.21
20 studies evaluated duration of viral RNA shedding based on disease severity. 13 of these studies reported longer duration of viral shedding in patients with severe illness than in those with non-severe illness,17, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34 whereas five studies in upper respiratory tract samples16, 18, 35, 36, 37 and one study in stool samples38 reported similar shedding durations according to disease severity. One study reported shorter duration of viral shedding in moderate to severe illness than in mild to moderate illness.39 Six studies compared viral shedding among individuals with severe illness versus non-severe illness:17, 23, 25, 26, 36, 37 five studies showed significantly longer duration of shedding among those with severe illness than among those with non-severe illness,17, 23, 25, 26, 36 and one study observed no difference37 (table 1 ).
Table 1.
Severity of illness and viral dynamics
| Classification of illness severity | Median (IQR*) duration of SARS-CoV-2 positivity in cohort, days | Viral dynamics in patients with severe illness vs those with non-severe illness | p value | |
|---|---|---|---|---|
| Chen et al (2020)25 | ICU vs non-ICU patients | 11 (95% CI 10–12) | Median time to viral clearance significantly longer in ICU vs non-ICU patients (HR 3·17, 95% CI 2·29–4·37) | Only HR provided |
| Chen et al (2020)26 | China CDC guideline (version 7) | 12 (8–16) | Shedding duration varies by severity: asymptomatic 6 days; mild 10 days; moderate 12 days; serious 14 days; critical 32 days | <0·0001 |
| Tan et al (2020)17 | China CDC guideline (version 6) | Nasopharyngeal swab: 12 (range 3–38); any sample: 22 (range 3–38) | Viral shedding significantly longer in patients with severe illness: any sample 23 days vs 20 days (note that nasopharyngeal swab 14 vs 11 days was non-significant) | 0·023 (any sample) |
| Xu et al (2020)36 | WHO criteria | 17 (13–32) | Higher proportion of patients with severe illness had shedding >15 days (34·2% vs 16·2%) | 0·049 |
| Yan et al (2020)37 | China CDC guideline (version 6) | 23 (18–32) | No difference in shedding duration (general illness 23 days vs severe illness 26 days vs critical illness 28 days) | 0·51 |
| Zheng et al (2020)23 | China CDC guideline (version 6) | Respiratory sample: 18 (13–29) | Shedding duration significantly longer in patients with severe illness (21 vs 14 days) in respiratory samples; no difference in shedding duration in stool or serum samples | 0·04 |
CDC=Center for Disease Control and Prevention. HR=hazard ratio. ICU=intensive care unit. SARS-CoV-2=severe acute respiratory syndrome coronavirus 2.
IQR unless otherwise stated.
All but one study40 that examined the effect of age on SARS-CoV-2 shedding identified an association between older age (older than 60 years) and prolonged viral RNA shedding.23, 24, 26, 31, 35, 36, 37, 41, 42, 43 Three studies identified age as an independent risk factor for delayed viral clearance.23, 24, 36 Male sex was also associated with prolonged shedding,23, 36, 44 and the association remained significant even when patients were stratified based on illness severity.23, 36 Corticosteroid treatment was associated with delayed viral clearance in four studies,31, 36, 45, 46 and one study that recruited 120 patients with critical illness found no difference between corticosteroid and control groups.47
A randomised, placebo-controlled trial of remdesivir in adults with severe COVID-19 found a similar decline in viral load over time in remdesivir and control groups, as well as similar proportions of patients with undetectable viral RNA at 28 days.102 In a phase 2, open-label study evaluating interferon beta-1b, lopinavir–ritonavir, and ribavirin, a shorter duration of viral shedding was seen with combination treatment than with the control.48 None of the antiviral regimens (chloroquine, oseltamivir, arbidol, and lopinavir–ritonavir) independently improved viral RNA clearance.26, 49 In a retrospective study of 284 patients, lopinavir–ritonavir use was associated with delayed viral clearance even after adjusting for confounders.26
12 studies reported viral load dynamics or duration of viral shedding among individuals with asymptomatic SARS-CoV-2 infection (table 2 ); two demonstrated lower viral loads among asymptomatic individuals than among symptomatic individuals,8, 50 and four found similar initial viral loads.13, 14, 51, 52 However, Chau and colleagues reported significantly lower viral load in asymptomatic individuals during the follow-up than in symptomatic individuals.51 Faster viral clearance was observed in asymptomatic individuals in five of six studies.13, 26, 51, 53, 54 The exception, Yongchen and colleagues, found longer shedding duration among asymptomatic cases, but the difference was not significant.34
Table 2.
SARS-CoV-2 viral dynamics in asymptomatic individuals compared with symptomatic individuals
| Median (IQR*) duration of SARS-CoV-2 positivity in asymptomatic individuals, days | Viral dynamics in asymptomatic vs symptomatic individuals | p value | |
|---|---|---|---|
| Arons et al (2020)52 | Not reported | No difference in viral load | Not reported |
| Chau et al (2020)51 | Not reported | Initial viral load similar; asymptomatic individuals had significantly lower viral load during follow-up and faster viral clearance than symptomatic individuals | 0·027 |
| Chen et al (2020)26 | 6 (4–10) | Significantly shorter duration of viral shedding among asymptomatic cases, with increasing shedding duration associated with increasing illness severity | <0·0001 |
| Han et al (2020)8 | Not reported | Symptomatic children had higher initial RNA load in nasopharyngeal swab specimens than asymptomatic children (9·01 vs 6·32 log10 copies per mL) | 0·048 |
| Hu et al (2020)53 | 6 (2–12) | Asymptomatic individuals had shorter duration of viral shedding compared with pre-symptomatic individuals (median duration of viral shedding was 6 days [2–12] vs 12 days [12–14]) | Not reported |
| Lavezzo et al (2020)14 | Not reported | No difference in viral load | p=0·62 (E gene); p=0·74 (RdRp gene) |
| Le et al (2020)57 | 9 | Not reported | Not applicable |
| Sakurai et al (2020)41 | 9 (6–11) | Not reported | Not applicable |
| Yang et al (2020)54 | 8 (3–12) | Significantly shorter duration of viral shedding from nasopharynx swabs was observed among asymptomatic vs symptomatic individuals | 0·001 |
| Yongchen et al (2020)34 | 18 (range 5–28) | Longer shedding duration among asymptomatic cases (median 18 days [range 5–28]) vs non-severe (10 days [2–21]) and severe (14 days [9–33]) cases | Not reported |
| Zhang et al (2020)13 | 9·6 | Initial viral load similar; viral clearance occurred earlier in the asymptomatic (9·6 days) and symptomatic individuals (9·7 days), vs pre-symptomatic group (13·6 days) | <0·05 |
| Zhou et al (2020)50 | Not reported | Significantly higher viral load in symptomatic (n=22) vs asymptomatic (n=9) individuals (median cycle threshold value 34·5 [IQR 37·5–39·5] vs 39·0 [32·2–37·0]), but duration of shedding was similar | Not reported |
SARS-CoV-2=severe acute respiratory syndrome coronavirus 2.
IQR, if available, unless otherwise stated.
We identified 11 studies that attempted to isolate live virus. All eight studies that attempted virus isolation in respiratory samples successfully cultured viable virus within the first week of illness.3, 9, 16, 52, 55, 56, 57, 58 No live virus was isolated from any respiratory samples taken after day 8 of symptoms in three studies,3, 55, 56 or beyond day 9 in two studies16, 52 despite persistently high viral RNA loads. One study demonstrated the highest probability of positive culture on day 3 of symptoms.55 Arons and colleagues cultured viable virus 6 days before typical symptom onset; however, onset of symptoms was unclear.52
The success of viral isolation correlated with viral load quantified by RT-PCR. No successful viral culture was obtained from samples with a viral load below 1 × 106 copies per mL in one study,3 cycle threshold values higher than 24 in another study,55 or higher than 34 in other studies,52, 56 with culture positivity declining with increasing cycle threshold values.56 Several other studies cultured live virus from RT-PCR-positive specimens; however, they did not correlate these results with viral load titres.9, 57, 58
One study reported the duration of viable virus shedding in respiratory samples; time to clearance from symptom onset was 3–12 days in upper respiratory tract samples and 5–13 days in lower respiratory tract samples, and no positive viral culture was obtained after day 4 in upper respiratory tract infection and day 8 in lower respiratory tract infection.3 Arons and colleagues cultured viable virus from the respiratory tract in one of three asymptomatic cases.52
Viral culture was successful in two of three RT-PCR-positive patients in one study, but the timepoints from symptom onset were not reported.59 Andersson and colleagues were unable to culture virus from 27 RT-PCR-positive serum samples.60
Of eight studies on SARS-CoV, none reported mean or median duration of viral shedding and thus were not eligible for quantitative analysis. The maximum duration of viral shedding reported was 8 weeks in upper respiratory tract,83, 84 52 days in lower respiratory tract,83, 85 6–7 weeks in serum,86 and 126 days in stool samples.83, 85, 87, 88, 89 Studies that evaluated SARS-CoV kinetics found low viral load in the initial days of illness, increasing after the first week of illness in upper respiratory tract samples, peaking at day 10,90 or days 12–14,87 and declining after weeks 3–4.84 High viral loads correlated with severity of illness and poor survival.84 Although Chen and colleagues identified an association between younger age and lower viral titres,84 Leong and colleagues found no difference.89 Viable SARS-CoV was isolated from stool and respiratory samples up to 4 weeks, and urine specimens up to day 36 from symptom onset.83, 86 All attempts to isolate virus from RT-PCR-positive stool specimens collected more than 6 weeks after disease onset failed.85 The isolation probability for stool samples was approximately five to ten times lower than for respiratory specimens.83
We identified 11 studies on MERS-CoV. Three studies (324 participants) reporting MERS-CoV shedding in the upper respiratory tract99, 100, 101 and four studies (93 participants) reporting MERS-CoV shedding in the lower respiratory tract91, 92, 96, 101 were included in the quantitative analysis. The mean shedding duration was 15·3 days (95% CI 11·6–19·0) in the upper respiratory tract and 16·3 days (13·8–18·9) in the lower respiratory tract (Figure 3, Figure 4 ). Only one study reported duration of viral shedding in serum with a maximum of 34 days.91 In a small study, mortality was higher in patients with viraemia (viral RNA in blood).92 In upper and lower respiratory tract specimens, prolonged shedding was associated with illness severity93, 94 and survival,95 with the shortest duration observed in asymptomatic individuals.93 Peak viral loads were observed between days 7 and 10, and higher viral loads were observed among patients with severe illness and fatal outcome.91, 93, 94, 96, 97 Differences in viral loads between survivors and fatal cases was more pronounced in the second week of illness (p=0·0006).97 The proportion of successful viable culture was 6% in respiratory samples, with a viral load value below 1 × 107 copies per mL.98
Figure 3.
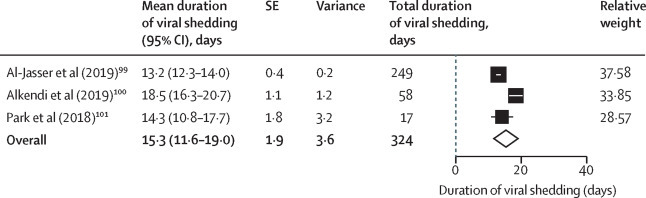
Pooled mean duration (days) of MERS-CoV shedding from the upper respiratory tract (random-effects model)
MERS-CoV=Middle East respiratory syndrome coronavirus.
Figure 4.
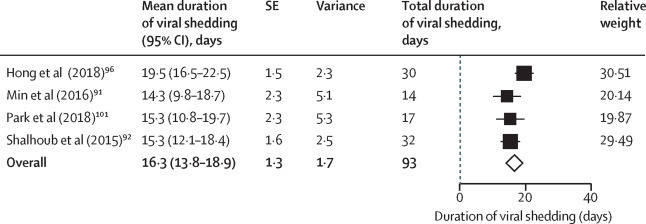
Pooled mean duration (days) of MERS-CoV shedding from the lower respiratory tract (random-effects model)
MERS-CoV=Middle East respiratory syndrome coronavirus.
All but 11 studies (six cohort studies, two cross-sectional studies, and one randomised controlled trial on SARS-CoV-2 and two cohort studies on MERS-CoV) were case series, the majority of which recruited non-consecutive patients and were therefore prone to possible selection bias (appendix p pp 9–13).
Discussion
This systematic review and meta-analysis provides comprehensive data on the viral dynamics of SARS-CoV-2, including the duration of RNA shedding and viable virus isolation. Our findings suggest that, although patients with SARS-CoV-2 infection might have prolonged RNA shedding of up to 83 days in upper respiratory tract infection, no live virus was isolated from culture beyond day 9 of symptoms despite persistently high viral RNA loads. This finding is supported by several studies demonstrating an association between viral load and viability of virus, with no successful culture from samples below a certain viral load threshold. These findings indicate that, in clinical practice, repeat testing might not be indicated to deem patients no longer infectious. Duration of infectiousness and subsequent isolation timelines could reflect viral load dynamics and could be counted from symptom onset for 10 days in non-severe cases.
SARS-CoV-2 viral load appeared to peak in the upper respiratory tract within the first week after symptom onset, and later in the lower respiratory tract. By contrast, the viral load of SARS-CoV peaked at days 10–14 of illness and that of MERS-CoV peaked at 7–10 days of illness. Combined with isolation of viable virus in respiratory samples primarily within the first week of illness, patients with SARS-CoV-2 infection are likely to be most infectious in the first week of illness, emphasising the importance of immediate isolation with symptom onset early in the course of illness. Several studies report viral load peaks during the prodromal phase of illness or at the time of symptom onset.2, 3, 4, 8, 15, 16, 17, 18, 19, 20, 21 providing a rationale for the efficient spread of SARS-CoV-2. This finding is supported by the observation in contact-tracing studies that the highest risk of transmission occurs very early in the disease course (a few days before and within the first 5 days after symptom onset).103, 104 Although modelling studies estimated potential viral load peak before symptom onset, we did not identify any study that confirms pre-symptomatic viral load peak.15
Similar to SARS-CoV, SARS-CoV-2 can be detected in stool samples for prolonged periods, with high viral loads detected even after 3 weeks of illness. In SARS-CoV, RNA prevalence in stool samples was high, with almost all studies reporting shedding in stools. Although viable SARS-CoV was isolated during up to 4 weeks of illness, faecal–oral transmission was not considered to be a primary driver of infection. By contrast, none of the studies in MERS-CoV reported duration of viral shedding in stool samples and RNA detection was low.97, 105 So far, only a few studies have demonstrated viable SARS-CoV-2 in stool samples.59, 106 Thus, the role of faecal shedding in viral transmission remains unclear.
Viral loads appear to be similar between asymptomatic and symptomatic individuals infected with SARS-CoV-2. Nevertheless, most studies demonstrate faster viral clearance among asymptomatic individuals than those who are symptomatic. This finding is in keeping with viral kinetics observed with other respiratory viruses such as influenza and MERS-CoV, in which people with asymptomatic infection have a shorter duration of viral shedding than symptomatic individuals.93, 107 However, data on the shedding of infectious virus in asymptomatic individuals are too scarce to quantify their transmission potential in order to inform policy on quarantine duration in the absence of testing.
To our knowledge, this is the first systematic review to comprehensively examine and compare SARS-CoV-2, SARS-CoV, and MERS-CoV viral dynamics, and the first meta-analysis of viral shedding duration. Our study has limitations. First, almost all patients in the included studies received a range of treatments, which might have modified the shedding dynamics. Second, our meta-analysis identified substantial study heterogeneity, probably due to differences in study population, follow-up, and management approaches. Furthermore, shedding duration is reported as median with IQR for most studies, but meta-analysis necessitates conversion to mean with SD.6 The validity of this conversion is based on the assumption that duration of viral shedding is normally distributed, which might not apply to some studies. Last, although there is probably a broad overlap, the true clinical window of infectious shedding might not entirely align with viral culture duration.
We identified a systematic review of SARS-CoV-2 viral load kinetics that included studies published up until May 12, 2020.108 This review included 26 case reports and 13 case series involving less than five individuals, which did not meet our eligibility criteria; these studies are prone to substantial selection bias, reporting atypical cases with prolonged viral shedding. Additionally, the review included studies that reported viral shedding duration from the time of hospital admission or initial PCR positivity. Furthermore, no meta-analysis of the duration of viral shedding was done.
This review provides detailed understanding about the evidence available so far on viral dynamics of SARS-CoV-2 and has implications for pandemic control strategies and infection control practices. Although SARS-CoV-2 RNA shedding can be prolonged in respiratory and stool samples, viable virus is short-lived, with culture success associated with viral load levels. Most studies detected the SARS-CoV-2 viral load peak within the first week of illness. These findings highlight that isolation practices should be commenced with the start of first symptoms, including mild and atypical symptoms that precede more typical COVID-19 symptoms. However, given potential delays in the isolation of patients, effective containment of SARS-CoV-2 might be challenging even with an early detection and isolation strategy.109
Data sharing
Data used in this study are available upon request from the corresponding author.
Acknowledgments
Acknowledgments
We thank Vicki Cormie at the University of St Andrews (Fife, UK) for assistance with the search and obtaining papers not readily accessible.
Contributors
MC was involved in conceptualisation, methodology, investigation, data curation, and writing of the original draft. MT was involved in investigation, data curation, and writing of the original draft. OL was involved in investigation, data curation, and review and editing of the manuscript. AEM was involved in formal analysis and writing of the original draft. JS was involved in investigation, data curation, and review and editing of the manuscript. AH was involved in conceptualisation, methodology, data curation, writing of the original draft, and supervision of the study.
Declaration of interests
We declare no competing interests.
Supplementary Material
References
- 1.Cevik M, Bamford CGG, Ho A. COVID-19 pandemic—a focused review for clinicians. Clin Microbiol Infect. 2020;26:842–847. doi: 10.1016/j.cmi.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zou L, Ruan F, Huang M. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wölfel R, Corman VM, Guggemos W. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 4.Kim ES, Chin BS, Kang CK. Clinical course and outcomes of patients with severe acute respiratory syndrome coronavirus 2 infection: a preliminary report of the first 28 patients from the Korean cohort study on COVID-19. J Korean Med Sci. 2020;35:e142. doi: 10.3346/jkms.2020.35.e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Joanna Briggs Institute The Joanna Briggs Institute Critical Appraisal tools for use in JBI systematic reviews. Checklist for case series. 2017. https://joannabriggs.org/sites/default/files/2019-05/JBI_Critical_Appraisal-Checklist_for_Case_Series2017_0.pdf
- 6.Wan X, Wang W, Liu J, Tong T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med Res Methodol. 2014;14:135. doi: 10.1186/1471-2288-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai J, Xu J, Lin D. A case series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin Infect Dis. 2020;71:1547–1551. doi: 10.1093/cid/ciaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han MS, Seong M-W, Kim N. Viral RNA load in mildly symptomatic and asymptomatic children with COVID-19, Seoul, South Korea. Emerg Infect Dis. 2020;26:2497–2499. doi: 10.3201/eid2610.202449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.L'Huillier AG, Torriani G, Pigny F, Kaiser L, Eckerle I. Culture-competent SARS-CoV-2 in nasopharynx of symptomatic neonates, children, and adolescents. Emerg Infect Dis. 2020;26:2494–2497. doi: 10.3201/eid2610.202403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan YP, Tan BY, Pan J, Wu J, Zeng SZ, Wei HY. Epidemiologic and clinical characteristics of 10 children with coronavirus disease 2019 in Changsha, China. J Clin Virol. 2020;127 doi: 10.1016/j.jcv.2020.104353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Q, Xing Y, Shi L. Epidemiological and clinical characteristics of children with coronavirus disease 2019. medRxiv. 2020 doi: 10.1101/2020.03.19.20027078. published online March 26. (preprint) [DOI] [Google Scholar]
- 12.Xu Y, Li X, Zhu B. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Xiao T, Wang Y. Early viral clearance and antibody kinetics of COVID-19 among asymptomatic carriers. medRxiv. 2020 doi: 10.1101/2020.04.28.20083139. published online May 2. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavezzo E, Franchin E, Ciavarella C. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo'. Nature. 2020;584:425–429. doi: 10.1038/s41586-020-2488-1. [DOI] [PubMed] [Google Scholar]
- 15.He X, Lau EHY, Wu P. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 16.Kujawski SA, Wong KK, Collins JP. Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat Med. 2020;26:861–868. doi: 10.1038/s41591-020-0877-5. [DOI] [PubMed] [Google Scholar]
- 17.Tan W, Lu Y, Zhang J. Viral kinetics and antibody responses in patients with COVID-19. medRxiv. 2020 doi: 10.1101/2020.03.24.20042382. published online March 26. (preprint) [DOI] [Google Scholar]
- 18.To KK-W, Tsang OT-Y, Leung W-S. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyllie AL, Fournier J, Casanovas-Massana A. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young BE, Ong SWX, Kalimuddin S. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323:1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J, Mao T, Li S. Long period dynamics of viral load and antibodies for SARS-CoV-2 infection: an observational cohort study. medRxiv. 2020 doi: 10.1101/2020.04.22.20071258. published online April 27. (preprint) [DOI] [Google Scholar]
- 22.Zhang N, Gong Y, Meng F, Bi Y, Yang P, Wang F. Virus shedding patterns in nasopharyngeal and fecal specimens of COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.03.28.20043059. published online March 30. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng S, Fan J, Yu F. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369 doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai Q, Huang D, Ou P. COVID-19 in a designated infectious diseases hospital outside Hubei Province, China. Allergy. 2020;75:1742–1752. doi: 10.1111/all.14309. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Qi T, Liu L. Clinical progression of patients with COVID-19 in Shanghai, China. J Infect. 2020;80:e1–e6. doi: 10.1016/j.jinf.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Zhu B, Hong W. Associations of clinical characteristics and treatment regimens with the duration of viral RNA shedding in patients with COVID-19. Int J Infect Dis. 2020;98:252–260. doi: 10.1016/j.ijid.2020.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Chen L, Deng Q. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J Med Virol. 2020;92:833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- 28.Fan L, Liu C, Li N. Medical treatment of 55 patients with COVID-19 from seven cities in northeast China who fully recovered: a single-center, retrospective, observational study. medRxiv. 2020 doi: 10.1101/2020.03.28.20045955. published online March 30. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang Z, Zhang Y, Hang C, Ai J, Li S, Zhang W. Comparisons of viral shedding time of SARS-CoV-2 of different samples in ICU and non-ICU patients. J Infect. 2020;81:147–178. doi: 10.1016/j.jinf.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Yan L-M, Wan L. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020;20:656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi J, Cheng C, Yu M. Systemic inflammatory cytokines associate with SARS-CoV-2 viral shedding time in Covid-19 inpatients. Res Sq. 2020 doi: 10.21203/rs.3.rs-31556/v1. published online May 29. (preprint) [DOI] [Google Scholar]
- 32.Tan L, Kang X, Ji X. Validation of predictors of disease severity and outcomes in COVID-19 patients: a descriptive and retrospective study. Med. 2020 doi: 10.1016/j.medj.2020.05.002. https://doi.org/10.1016%2Fj.medj.2020.05.002 published online May 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou F, Yu T, Du R. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yongchen Z, Shen H, Wang X. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerg Microbes Infect. 2020;9:833–836. doi: 10.1080/22221751.2020.1756699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu X, Xing Y, Jia J. Factors associated with negative conversion of viral RNA in patients hospitalized with COVID-19. Sci Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu K, Chen Y, Yuan J. Factors associated with prolonged viral RNA shedding in patients with coronavirus disease 2019 (COVID-19) Clin Infect Dis. 2020;71:799–806. doi: 10.1093/cid/ciaa351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan D, Liu X-Y, Zhu YN. Factors associated with prolonged viral shedding and impact of lopinavir/ritonavir treatment in hospitalised non-critically ill patients with SARS-CoV-2 infection. Eur Respir J. 2020;56 doi: 10.1183/13993003.00799-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y, Guo C, Tang L. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian D, Wang L, Wang X. Clinical research and factors associated with prolonged duration of viral shedding in patients with COVID-19. Res Sq. 2020 doi: 10.21203/rs.3.rs-29818/v1. published online June 1. (preprint) [DOI] [Google Scholar]
- 40.Zhou B, She J, Wang Y, Ma X. The duration of viral shedding of discharged patients with severe COVID-19. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa451. published online April 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakurai A, Sasaki T, Kato S. Natural history of asymptomatic SARS-CoV-2 infection. N Engl J Med. 2020;383:885–886. doi: 10.1056/NEJMc2013020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talmy T, Tsur A, Shabtay O. Duration of SARS-CoV-2 detection in Israel Defense Forces soldiers with mild COVID-19. J Med Virol. 2020 doi: 10.1002/jmv.26374. published online July 30. [DOI] [PubMed] [Google Scholar]
- 43.Xiao AT, Tong YX, Zhang S. Profile of RT-PCR for SARS-CoV-2: a preliminary study from 56 COVID-19 patients. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa460. published online April 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shastri A, Wheat J, Agrawal S. Delayed clearance of SARS-CoV2 in male compared to female patients: high ACE2 expression in testes suggests possible existence of gender-specific viral reservoirs. medRxiv. 2020 doi: 10.1101/2020.04.16.20060566. published online April 17. (preprint) [DOI] [Google Scholar]
- 45.Ling Y, Xu S-B, Lin Y-X. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J. 2020;133:1039–1043. doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zha L, Li S, Pan L. Corticosteroid treatment of patients with coronavirus disease 2019 (COVID-19) Med J Aust. 2020;212:416–420. doi: 10.5694/mja2.50577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang M, Chen P, He M. Corticosteroid treatment in critically ill patients with COVID-19: a retrospective cohort study. Res Sq. 2020 doi: 10.21203/rs.3.rs-27386/v1. published online May 11. (preprint) [DOI] [Google Scholar]
- 48.Hung IFN, Lung KC, Tso EYK. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang H, Guan L, Yang Y. Chloroquine, arbidol (umifenovir) or lopinavir/ritonavir as the antiviral monotherapy for COVID-19 patients: a retrospective cohort study. Res Sq. 2020 doi: 10.21203/rs.3.rs-24667/v1. published online April 24. (preprint) [DOI] [Google Scholar]
- 50.Zhou R, Li F, Chen F. Viral dynamics in asymptomatic patients with COVID-19. Int J Infect Dis. 2020;96:288–290. doi: 10.1016/j.ijid.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chau NVV, Thanh Lam V, Thanh Dung N. The natural history and transmission potential of asymptomatic SARS-CoV-2 infection. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa711. published online June 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arons MM, Hatfield KM, Reddy SC. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med. 2020;382:2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu Z, Song C, Xu C. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci. 2020;63:706–711. doi: 10.1007/s11427-020-1661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang R, Gui X, Xiong Y. Comparison of clinical characteristics of patients with asymptomatic vs symptomatic coronavirus disease 2019 in Wuhan, China. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bullard J, Dust K, Funk D. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa638. published online May 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.La Scola B, Le Bideau M, Andreani J. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le TQM, Takemura T, Moi ML. Severe acute respiratory syndrome coronavirus 2 shedding by travelers, Vietnam, 2020. Emerg Infect Dis. 2020;26:1624–1626. doi: 10.3201/eid2607.200591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.To KK-W, Tsang OT-Y, Yip CC-Y. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71:841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiao F, Sun J, Xu Y. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg Infect Dis J. 2020;26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andersson M, Arancibia-Carcamo CV, Auckland K. SARS-CoV-2 RNA detected in blood samples from patients with COVID-19 is not associated with infectious virus. medRxiv. 2020 doi: 10.1101/2020.05.21.20105486. published online June 17. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang D, Mo G, Yuan X. Time kinetics of viral clearance and resolution of symptoms in novel coronavirus infection. Am J Respir Crit Care Med. 2020;201:1150–1152. doi: 10.1164/rccm.202003-0524LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen X, Ling J, Mo P. Restoration of leukomonocyte counts is associated with viral clearance in COVID-19 hospitalized patients. medRxiv. 2020 doi: 10.1101/2020.03.03.20030437. published online March 6. (preprint). [DOI] [Google Scholar]
- 63.Corman VM, Rabenau HF, Adams O. SARS-CoV-2 asymptomatic and symptomatic patients and risk for transfusion transmission. Transfusion. 2020;60:1119–1122. doi: 10.1111/trf.15841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fu S, Fu X, Song Y. Virologic and clinical characteristics for prognosis of severe COVID-19: a retrospective observational study in Wuhan, China. medRxiv. 2020 doi: 10.1101/2020.04.03.20051763. published online April 6. (preprint). [DOI] [Google Scholar]
- 65.Huang L, Chen Z, Ni L. Impact of angiotensin-converting enzyme inhibitors and angiotensin receptor blockers on inflammatory responses and viral clearance in COVID-19 patients: a multicenter retrospective cohort study. Res Sq. 2020 doi: 10.21203/rs.3.rs-27366/v1. published online May 8. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li N, Wang X, Lv T. Prolonged SARS-CoV-2 RNA shedding: not a rare phenomenon. J Med Virol. 2020;92:2286–2287. doi: 10.1002/jmv.25952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu L, Liu W, Zheng Y. A preliminary study on serological assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 238 admitted hospital patients. Microbes Infect. 2020;22:206–211. doi: 10.1016/j.micinf.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lo IL, Lio CF, Cheong HH. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int J Biol Sci. 2020;16:1698–1707. doi: 10.7150/ijbs.45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lou B, Li T-D, Zheng S-F. Serology characteristics of SARS-CoV-2 infection after exposure and post-symptom onset. Eur Respir J. 2020;56 doi: 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pongpirul WA, Mott JA, Woodring JV. Clinical characteristics of patients hospitalized with coronavirus disease, Thailand. Emerg Infect Dis. 2020;26:1580–1585. doi: 10.3201/eid2607.200598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qian GQ, Chen XQ, Lv DF. Duration of SARS-CoV-2 viral shedding during COVID-19 infection. Infect Dis. 2020;52:511–512. doi: 10.1080/23744235.2020.1748705. [DOI] [PubMed] [Google Scholar]
- 72.Quan W, Zheng Q, Tian J. No SARS-CoV-2 in expressed prostatic secretion of patients with coronavirus disease 2019: a descriptive multicentre study in China. medRxiv. 2020 doi: 10.1101/2020.03.26.20044198. published online March 30. (preprint) [DOI] [Google Scholar]
- 73.Seah IYJ, Anderson DE, Kang AEZ. Assessing viral shedding and infectivity of tears in coronavirus disease 2019 (COVID-19) patients. Ophthalmology. 2020;127:977–979. doi: 10.1016/j.ophtha.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Song C, Wang Y, Li W. Detection of 2019 novel coronavirus in semen and testicular biopsy specimen of COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.03.31.20042333. published online April 10. (preprint). [DOI] [Google Scholar]
- 75.Song R, Han B, Song M. Clinical and epidemiological features of COVID-19 family clusters in Beijing, China. J Infect. 2020;81:e26–e30. doi: 10.1016/j.jinf.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tu Y-H, Wei Y-Y, Zhang D-W, Chen C-S, Hu X-W, Fei G. Analysis of factors affected the SARS-CoV-2 viral shedding time of COVID-19 patients in Anhui, China: a retrospective study. Res Sq. 2020 doi: 10.21203/rs.3.rs-20954/v1. published online April 6. (preprint) [DOI] [Google Scholar]
- 77.Wang L, Gao Y-h, Lou L-L, Zhang G-J. The clinical dynamics of 18 cases of COVID-19 outside of Wuhan, China. Eur Respir J. 2020;55 doi: 10.1183/13993003.00398-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang S, Tu J, Sheng Y. Clinical characteristics and fecal-oral transmission potential of patients with COVID-19. medRxiv. 2020 doi: 10.1101/2020.05.02.20089094. published online May 6. (preprint) [DOI] [Google Scholar]
- 79.Wu B, Lei Z-Y, Wu K-L. Epidemiological and clinical features of imported and local patients with coronavirus disease 2019 (COVID-19) in Hainan, China. SSRN. 2020 doi: 10.2139/ssrn.3555222. published online March 24. (preprint) [DOI] [Google Scholar]
- 80.Xu l, Zhang X, Song W. Conjunctival polymerase chain reaction-tests of 2019 novel coronavirus in patients in Shenyang, China. medRxiv. 2020 doi: 10.1101/2020.02.23.20024935. published online Feb 25. (preprint) [DOI] [Google Scholar]
- 81.Yang Y, Yang M, Shen C. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv. 2020 doi: 10.1101/2020.02.11.20021493. published online Feb 17. (preprint) [DOI] [Google Scholar]
- 82.Zhu L, Gong N, Liu B. Coronavirus disease 2019 pneumonia in immunosuppressed renal transplant recipients: a summary of 10 confirmed cases in Wuhan, China. Eur Urol. 2020;18:18. doi: 10.1016/j.eururo.2020.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chan PK, To WK, Ng KC. Laboratory diagnosis of SARS. Emerg Infect Dis. 2004;10:825–831. doi: 10.3201/eid1005.030682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen WJ, Yang JY, Lin JH. Nasopharyngeal shedding of severe acute respiratory syndrome-associated coronavirus is associated with genetic polymorphisms. Clin Infect Dis. 2006;42:1561–1569. doi: 10.1086/503843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu W, Tang F, Fontanet A. Long-term SARS coronavirus excretion from patient cohort, China. Emerg Infect Dis. 2004;10:1841–1843. doi: 10.3201/eid1010.040297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu D, Zhang Z, Jin L. Persistent shedding of viable SARS-CoV in urine and stool of SARS patients during the convalescent phase. Eur J Clin Microbiol Infect Dis. 2005;24:165–171. doi: 10.1007/s10096-005-1299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cheng PK, Wong DA, Tong LK. Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet. 2004;363:1699–1700. doi: 10.1016/S0140-6736(04)16255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kwan BC, Leung CB, Szeto CC. Severe acute respiratory syndrome in dialysis patients. J Am Soc Nephrol. 2004;15:1883–1888. doi: 10.1097/01.asn.0000131522.16404.1f. [DOI] [PubMed] [Google Scholar]
- 89.Leong HN, Chan KP, Khan AS. Virus-specific RNA and antibody from convalescent-phase SARS patients discharged from hospital. Emerg Infect Dis. 2004;10:1745–1750. doi: 10.3201/eid1010.040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peiris JSM, Chu CM, Cheng VCC. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Min CK, Cheon S, Ha NY. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity. Sci Rep. 2016;6 doi: 10.1038/srep25359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shalhoub S, Farahat F, Al-Jiffri A. IFN-α2a or IFN-β1a in combination with ribavirin to treat Middle East respiratory syndrome coronavirus pneumonia: a retrospective study. J Antimicrob Chemother. 2015;70:2129–2132. doi: 10.1093/jac/dkv085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Al Hosani FI, Pringle K, Al Mulla M. Response to emergence of Middle East respiratory syndrome coronavirus, Abu Dhabi, United Arab Emirates, 2013–2014. Emerg Infect Dis. 2016;22:1162–1168. doi: 10.3201/eid2207.160040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Oh MD, Park WB, Choe PG. Viral load kinetics of MERS coronavirus infection. N Engl J Med. 2016;375:1303–1305. doi: 10.1056/NEJMc1511695. [DOI] [PubMed] [Google Scholar]
- 95.Arabi YM, Al-Omari A, Mandourah Y. Critically ill patients with the Middle East respiratory syndrome: a multicenter retrospective cohort study. Crit Care Med. 2017;45:1683–1695. doi: 10.1097/CCM.0000000000002621. [DOI] [PubMed] [Google Scholar]
- 96.Hong KH, Choi JP, Hong SH. Predictors of mortality in Middle East respiratory syndrome (MERS) Thorax. 2018;73:286–289. doi: 10.1136/thoraxjnl-2016-209313. [DOI] [PubMed] [Google Scholar]
- 97.Corman VM, Albarrak AM, Omrani AS. Viral shedding and antibody response in 37 patients with Middle East respiratory syndrome coronavirus infection. Clin Infect Dis. 2016;62:477–483. doi: 10.1093/cid/civ951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Muth D, Corman VM, Meyer B. Infectious Middle East respiratory syndrome coronavirus excretion and serotype variability based on live virus isolates from patients in Saudi Arabia. J Clin Microbiol. 2015;53:2951–2955. doi: 10.1128/JCM.01368-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Al-Jasser FS, Nouh RM, Youssef RM. Epidemiology and predictors of survival of MERS-CoV infections in Riyadh region, 2014–2015. J Infect Public Health. 2019;12:171–177. doi: 10.1016/j.jiph.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Alkendi F, Nair SC, Hashmey R. Descriptive epidemiology, clinical characteristics and outcomes for Middle East respiratory syndrome coronavirus (MERS-CoV) infected patients in AlAin – Abu Dhabi Emirate. J Infect Public Health. 2019;12:137. [Google Scholar]
- 101.Park WB, Poon LLM, Choi SJ. Replicative virus shedding in the respiratory tract of patients with Middle East respiratory syndrome coronavirus infection. Int J Infect Dis. 2018;72:8–10. doi: 10.1016/j.ijid.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang Y, Zhang D, Du G. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Y, Tian H, Zhang L. Reduction of secondary transmission of SARS-CoV-2 in households by face mask use, disinfection and social distancing: a cohort study in Beijing, China. BMJ Glob Health. 2020;5 doi: 10.1136/bmjgh-2020-002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cheng H-Y, Jian S-W, Liu D-P, Ng TC, Huang WT, Lin HH. Contact tracing assessment of COVID-19 transmission dynamics in Taiwan and risk at different exposure periods before and after symptom onset. JAMA Intern Med. 2020;180:1156–1163. doi: 10.1001/jamainternmed.2020.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Memish A. Viral shedding and antibody response in MERS. Vox Sang. 2016;111(suppl 1):48–49. [Google Scholar]
- 106.Wang W, Xu Y, Gao R. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ip DKM, Lau LLH, Leung NHL. Viral shedding and transmission potential of asymptomatic and paucisymptomatic influenza virus infections in the community. Clin Infect Dis. 2017;64:736–742. doi: 10.1093/cid/ciw841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Walsh KA, Jordan K, Clyne B. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect. 2020;81:357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kretzschmar ME, Rozhnova G, Bootsma MCJ, van Boven M, van de Wijgert JHHM, Bonten MJM. Impact of delays on effectiveness of contact tracing strategies for COVID-19: a modelling study. Lancet Public Health. 2020;5:e452–e459. doi: 10.1016/S2468-2667(20)30157-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used in this study are available upon request from the corresponding author.


