Abstract
SARS-COV-2 infection represents the greatest pandemic of the world, counting daily increasing number of subjects positive to the virus and, sadly, increasing number of deaths. Current studies reported that the cytokine/chemokine network is crucial in the onset and maintenance of the “cytokine storm”, the event occurring in those patients in whom the progression of COVID-19 will progress, in most cases, to a very severe and potentially threatening disease. Detecting a possible “immune signature” in patients, as assessed by chemokines status in patients with COVID-19, could be helpful for individual risk stratification for developing a more or less severe clinical course of the disease. The present review is specifically aimed at overviewing current evidences provided by in vitro and in vivo studies addressing the issue of which chemokines seems to be involved, at least at present, in COVID-19. Currently available experimental and clinical studies regarding those chemokines more deeply studied in COVID-19, with a specific focus on their role in the cytokine storm and ultimately with their ability to predict the clinical course of the disease, will be taken into account. Moreover, similarities and differences between chemokines and cytokines, which both contribute to the onset of the pro-inflammatory loop characterizing SARS-COV-2 infection, will be briefly discussed. Future studies will rapidly accumulate in the next months and their results will hopefully provide more insights as to the complex physiopathology of COVID-19-related cytokine storm. This will likely make the present review somehow “dated” in a short time, but still the present review provides an overview of the scenario of the current knowledge on this topic.
Abbreviations: SARS-COV-2, severe acute respiratory syndrome coronavirus 2; ACE-2, angiotensin-converting enzyme-2; Covid-19, coronavirus disease 2019; CS, cytokine storm; IL-6, interleukin-6; CXCR3, CXC-chemokine receptor 3; TNFα, tumor-necrosis-factor- α; IL-1, interleukin-1; ATP, adenosine triphosphate; IFN-γ, interferon-γ; IL1β, interleukin-1 β; IL-17, interleukin-17; Th17, T-Helper-17; RNA-Seq, RNA sequencing; MIS-C, multisystem inflammatory syndrome in children; MERS-CoV, middle east respiratory syndrome coronavirus; JAK, Janus kinase; ssGSEA, single sample gene set enrichment analysis; ARDS, acute respiratory distress syndrome; GSCF, granulocyte colony-stimulating factor; ICU, intensive care unit; HIV-1-2, human immunodeficiency virus-1-2; PBMC, peripheral blood mononuclear cell; BALF, bronchoalveolar lavage fluid; scRNA-seq, single-cell RNA sequencing; CXCL1, C-X-C Motif Chemokine Ligand 1; CXCL8, C-X-C Motif Chemokine Ligand 8; CXCL9, C-X-C Motif Chemokine Ligand 9; CXCL10, C-X-C Motif Chemokine Ligand 10; CCL2, C-C Motif Chemokine Ligand 2; CCL3, C-C Motif Chemokine Ligand 3; CCL5, C-C Motif Chemokine Ligand 5; CCL19, C-C Motif Chemokine Ligand 19; CCL20, C-C Motif Chemokine Ligand 20
Keywords: COVID-19-coronavirus, Chemokines, CXCL10, CXCL8 - cytokine storm, Chemokine receptor
1. SARS-COV-2 infection and the “cytokine storm”
The dramatic events due to the devastating epidemic caused by SARS-COV-2 have prompted researchers to make a common effort in the search for a possible solution to counter this new virus. The SARS-COV-2 infection involves 220 countries, areas or territories all over the world with a number of peoples positive to the virus >70.000.000 and more than 1.600.000 deaths [1].
Following the demonstration of the pathological role of the so called “cytokine storm”, several studies were designed with the aim of investigating whether specific chemokines could play a role in driving COVID-19 progression. These type of studies were aimed not only at identifying possible targets for treatment of the disease but also at detecting a possible “immune signature” in patients with COVID-19. Indeed, COVID-19 is a very heterogeneous disease, characterized by a clinical course which may range from a rather free of symptoms condition to fatal events. Thus, it is evident that the identification of a specific “immune signature” (i.e. high or low concentrations of one or more specific chemokine) of COVID-19 patients could potentially represent an helpful clinical tool for early identification, already in the early stages of the infection, of patients more or less prone to develop a severe clinical condition.
The present review will be aimed at providing an overview of the currently available experimental and clinical studies, addressing the issue of which chemokines are more deeply involved in the pathogenesis of the cytokine storm, potentially driving a more or less severe clinical course. A specific focus will be to highlight, on the basis of the rapidly growing number of studies, which are the most relevant chemokines sustaining the inflammatory consequences of the SARS-COV-2 infection-related cytokine storm and how their concentrations may reflect the degree of severity of COVID-19 as well as how their changes parallel the clinical outcome.
2. Chemokines involved in SARS-COV-2 infection
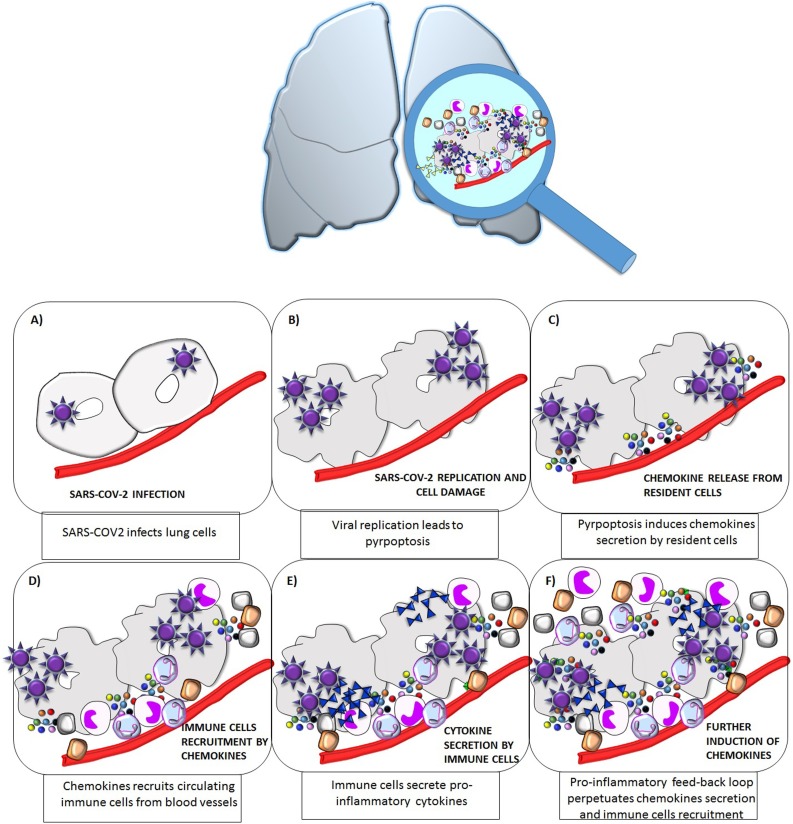
Protective immunity to viruses is dependent upon the activation and interplay between cytokines and chemokines to enhance/regulate innate or adaptive (or both) effector functions. In vitro and in vivo evidences, demonstrated that the chemokine milieu induced by one pathogen, by specifically recruiting T cells in the infected tissue, plays a crucial role in determining the nature of the immune response, [2,3]. The pro-inflammatory feed-back loop resulting from SARS-COV-2 infection generate a massive production of chemokines, which orchestrate the immune cells infiltration and the further secretion of both chemokine and cytokines resulting in the cytokine storm (Fig. 1 ). The particular importance of chemokines in COVID-19 was highlighted by the analysis of bronchoalveolar lavage fluid (BALF). Xiong et al, found that peripheral blood mononuclear cells (PBMCc) isolated from the BALF of COVID-19 patients, over-expressed the genes encoding for several chemokines, including CCL2, CXCL10, CCL3 and CCL4 [5]. Similarly, Zhou et al as well as Liao et al, reported, that lung macrophages from patients with severe COVID-19 infection, showed over-expression of the genes encoding for CCL2, CCL3, CCL4 and CCL7 (and consequently of the cytokines IL1β, IL6, TNFα) [6,7]. Moreover, Chua et al., reported an increase in activated immune cells, including inflammatory macrophages in bronchial samples from patients with COVID-19. These macrophages overexpressed the genes encoding for several chemokines, including CXCL10, CXCL8, CCL2, CCL3, CCL20, CXCL1, CXCL3, (and cytokines IL1β and TNF) [8]. This overexpression was found at higher magnitude in those patients with critical as compared to those with moderate disease [8]. Similarly, Chi et al., recently showed that patients with severe COVID-19 had higher serum levels of CXCL10, CCL2, CCL7, CXCL9, and CCL3 compared with mild cases, and higher levels of CCL7, CXCL9, and CCL3 compared with moderate cases [9]. More importantly, Lucas et al., demonstrated that a specific panel of chemokines would be responsible for monocyte recruitment being their circulating levels positively correlated with viral load only in patients with severe disease. Furthermore CCL1, CCL2, CCL4, CCL8, CCL21, CXCL9 were elevated already in the early phase of infection in those patients who ultimately died of COVID-19 [10]. Finally, transcript profiles obtained from lung autopsy samples, showed a significant upregulation of chemokines (CCL2, CXCL5, and CXCL6), in samples from COVID-19 patients as compared to samples from healthy control [11]. This finding was also confirmed in SARS-CoV-2-infected pancreatic endocrine cells compared to mock infected cells. Taken together the above results, it was consistently reported that many chemokines are involved in COVID-19 potentially influencing its progression and risk of mortality [[11], [12], [13]]. However, only few of these chemokines were object of studies specifically aimed at a better characterization of their role and at providing an explanation of their possible clinical utility in the management of the disease. In particular, there were 9 chemokines which, according to current knowledge, seem to play a role in SARS-COV-2 infection. Here follows a description of the more recent evidence of the involvement of each chemokine in COVID-19.
Fig. 1.
Schematic representation of the pro-inflammatory feed-back loop resulting from SARS−COV-2 infection in lung cells.
SARS−COV-2 enters lung cells by binding to the ACE-2 receptor (A); Intracellular viral replication is followed by cell stress/damage, inducing pyro-ptosis, which in turn produces several damage- associated-molecular-patterns (i.e. ATP, oligomers and nucleic acids) (B); As a consequence, resident lung cells start secreting chemokines (C); which will, in turn recruit immune cells expressing the specific chemokine receptor, through a multistep process involving adherence and migration across the endothelium, trafficking through the interstitium, and finally moving the site of the infection (D); Infiltrating immune cells start secreting pro-inflammatory cytokines (E); the onset and maintenance of this pro-inflammatory feed-back loop will result in mainteinance of chemokines secretion which will perpetuate the recruitment of infiltrating immune cells.
The above described events will ultimately result in the cytokine storm which will subsequently spread to other organs, eventually leading to multi-organ failure.
3. Hints for reading the following section
For the purpose of the present review, we followed in the subsequent paragraphs a “family order”, in that, starting from CXC family of chemokines, each chemokine was listed in ascending numerical order. In order to show a rapid link between a given chemokine and the studies evaluating it, Table 1 was provided. In particular, chemokines less deeply investigated (single study and/or evaluated within a wide panel of other chemokines) as well as those chemokines for which more data are available are listed in Table 1.
Table 1.
A summary of chemokines mainly involved in COVID-19 linked with respective studies.
| CHEMOKINE | STUDIES | REFERENCE |
|---|---|---|
| CXC FAMILY | ||
| CXCL1 | Chua et al.; Ghebremedhin et al. | [7,15] |
| CXCL3 | Chua et al. | [7] |
| CXCL5 | Blanco-Melo et al.; Vanderheiden et al. | [10,18] |
| CXCL6 | Blanco-Melo et al.; Vanderheiden et al. | [10,18] |
| CXCL8 | Chua et al. ; Vanderheiden et al. ; Liu et al. ; Li et al. ; Tincati et al; Diorio et al. ; Del Valle et al., Moratto et al. | [7,[18], [19], [20],[22], [23], [24],28] |
| CXCL9 | Chi et al, Lucas et al Moratto et al. Chen et al.;, Kwon et al. | [8,9,28,36,45] |
| CXCL10 | Xiong et al.; Chua et al.; Chi et al., Li et al., Huang et al.; Tincati et al., Moratto et al.; Yang et al. ; Zhang et al. ; Zhang et al. ; Hue et al. ; Chen et al.; Blot et al., Kwon et al., Yao et al., Blot et al. | [4,7,8,[20], [21], [22],28,[32], [33], [34], [35], [36], [37],45,54,65] |
| CCL13 | Lucas et al | [9] |
| CC FAMILY | ||
| CCL1 | Lucas et al | [9] |
| CCL2 | Xiong et al. ; Zhou et al.; Liao et al.; Chua et al. ; Chi et al., Lucas et al., Blanco-Melo et al. ; Li et al.; Huang et al. ; Tincati et al., Moratto et al. ; Zhang et al. ; Chen et al.; Abers et al.;Kwon et al., Sierra et al., Yao et al. | [[4], [5], [6], [7], [8], [9], [10],[20], [21], [22],28,34,36,[44], [45], [46],54] |
| CCL3 | Xiong et al.; Zhou et al. ; Liao et al. ; Chua et al., Chi et al.,Chen et al. ; Sierra et al.; Gruber et al. ; Yao et al. | [[4], [5], [6], [7], [8],36,46,50,54] |
| CCL4 | Xiong et al. ; Zhou et al. ; Liao et al. ; Lucas et al.; Gruber et al. | [[4], [5], [6],9,50] |
| CCL5 | Lucas et al; Li et al. ; Moratto et al. ; Chen et al. ; Yao et al., | [9,20,28,36,54] |
| CCL7 | Zhou et al. ; Liao et al. ; Chi et al; Yang et al. | [5,6,8,32] |
| CCL8 | Lucas et al | [9] |
| CCL15 | Lucas et al | [9] |
| CCL10 | Yao et al. | [54] |
| CCL19 | Balnis et al. | [60] |
| CCL20 | Chua et al. ; Huang et al. ; Gruber et al., Blot et al. | [7,21,50,65] |
| CCL21 | Lucas et al | [9] |
| CCL22 | Lucas et al | [9] |
| CCL27 | Lucas et al | [9] |
| CCL28 | Gruber et al. | [50] |
In order to facilitate reading, all chemokines will be listed in the heading of the paragraph with both old and new nomenclature, while only the new nomenclature will be used along the text. For clarity purpose, data from in vitro studies will anticipate those obtained by in vivo studies, unless specifically requested. In particular, since most studies evaluated more than one chemokine, the results will be reported with more details in the sub-paragraph of the specific chemokine that was found to be either more extensively evaluated or more relevant. All studies evaluating a specific chemokine are listed in Table 1, also including those chemokines less deeply investigated (single study and/or evaluated within a wide panel of other chemokines).
4. Chemokines belonging to the CXC family
4.1. CXCL1/GROα (Growth-regulated oncogene α)
CXCL1 (C-X-C Motif Chemokine Ligand 1) is a member of the CXC subfamily of chemokines. CXCL1 binds to the G-protein coupled receptor, CXC receptor 2, and it is considered to be a signalling growth factor [14,15]. CXCL1 is a known chemoattractant for neutrophils, playing a role in the inflammatory process. An abnormal expression and secretion of this chemokine is also associated with the growth and progression of some types of tumours [14]. There are few literature data regarding the role of this chemokine in SARS-COV-2 infection. This chemokine was identified, among others, to be the one of the more abundantly expressed by macrophages involved in SARS-COV2 infection, especially in more severe cases [8]. In addition, in vitro evidences, obtained in an established model of lung fibrosis induced by SARS-COV-2, demonstrated that the reduction of CXCL1 and other inflammatory markers in the lung by the peptide RP-832c would parallel the reduction of lung fibrosis [16].
4.2. CXCL8/IL-8 (Interleukin-8)
CXCL8 is a member of the CXC chemokine family commonly referred to as interleukin-8 (IL-8) according to the old nomenclature. CXCL8 is secreted by mononuclear macrophages, neutrophils, eosinophils, T lymphocytes, epithelial cells, and fibroblasts. It functions as a chemotactic factor by recruiting neutrophils to the site of infection. CXCL8 was indeed the first chemokine identified. More recently, CXCL8 became increasingly studied in different types of human cancer owing to its well established pro-tumorigenic effects [17,18]. CXCL8 seems to play an important role also in COVID-19 disease. In vitro evidence, by RNA-Seq data from human bronchial airway epithelial cells, showed that SARS-CoV-2 infection upregulates the production CXCL8 and other factors from both the apical and basolateral cell surface [19]. Moving to in vivo results, a retrospective study performed on demographic data, epidemiological history, clinical manifestations, and laboratory findings of patients with COVID-19, demonstrated that, together with other inflammatory molecules, the circulating levels of CXCL8 were significantly higher in patients bearing a more severe disease as compared to those with milder clinical symptoms [20]. These data suggested a positive correlation between CXCL8 levels and the degree of COVID-19 severity. Further support to this concept stems from a study on 69 patients with COVID-19 showing that serum CXCL8 (and IL-6) levels were, among a panel of other cytokines, remarkably higher in the severe group compared with the non-severe group. In particular, high CXCL8 levels persisted until hospital day-5, being associated with disease progression [21]. The Authors, suggested that, the neutrophilia, frequently occurring in severe COVID-19, might be ascribed to the abnormal elevation of CXCL8 and granulocyte-macrophage colony-stimulating factor [21,22]. In a subsequent study, a direct comparison between patients with severe versus moderate disease found that higher levels of inflammatory chemokines including MCP-1/CCL2, IP-10/CXCL10, IL-8/CXCL8, are found in patients with critical illness, correlating with both neutrophilia and increased circulating pro-inflammatory CD14+ CD16++ monocytes [23].
Moreover, Diorio et al, found an increase in markers of endothelial dysfunction in both children affected by Multisystem inflammatory syndrome (MIS-C) or severe COVID-19. Indeed elevated sC5b-9 (a marker of endothelial dysfunction) correlated with increased CXCL8 (as well as IL-6 and TNF-α) levels, suggesting a link between CXCL8 levels and innate immunity dysfunction commonly observed in these patients [24]. In addition, elevated serum concentrations of CXCL8 (together with other factors) at admission were also found to be strong and independent predictors of survival in hospitalized patients with COVID-19 [25].
Finally, it seems worth remembering that BMS-986253, a monoclonal antibody inhibiting CXCL8, previously tested in a Phase I clinical trial on cancer patients [26] is currently object of an ongoing Phase 2 clinical trial in patients affected by severe COVID-19 (NCT04347226).
4.3. CXCL9/MIG (Monokine induced by interferon-γ)
CXCL9 (C-X-C Motif Chemokine Ligand 9) is a CXC chemokine with chemoattractant activity for lymphocytes but not for neutrophils binding to C-X-C motif chemokine 3. This chemokine is involved in several inflammatory processes being (as CXCL10) chemoattractant for immune system cells of Th-1 phenotype [27,28]. This chemokine was found to be involved in COVID-19 disease being positively correlated to the age of the patients. Moratto et al., measured a panel of chemokines in plasma samples from 14 children and 9 adults with COVID-19 and in respective age-matched control groups. Briefly, adult patients with COVID-19 displayed higher levels of CXCL9 than children with COVID-19, while overlapping CXCL9 concentrations were observed between adult and children control subjects. Interestingly enough, the adults involved in the study also displayed a prominent Th1 polarization of immune response against SARS-CoV2 suggesting that prolonged T cell activation in adults with COVID-19 might result in Th1 polarization and possibly in pulmonary tissue damage by activated T cells [29].
Of note, this age-dependent difference was not observed for plasma levels of other chemokines including CCL5, CXCL10, CCL2, and CXCL8. Although of potential interest, the clinical significance of this age-related behaviour, remains at present yet to be fully elucidated.
4.4. CXCL10/IP-10 (interferon γ-inducible protein 10 kDa)
CXCL10 belongs to the CXC family of chemokines containing a single and variable amino acid between the two first of four highly conserved cysteine residues. This chemokine binds to CXCR3 receptor through which it exerts multiple functions [30]. CXCL10 is secreted after induction by IFN-γ by a wide variety of human cell types including immune cells and resident cells. High level of CXCL10 in peripheral fluids represent a marker of host immune response, specifically of Th1 orientated T-cells [31]. CXCL10 is at present the most studied chemokine within SARS-COV-2 infection, being consistently identified by several studies as the chemokine playing the major role in the SARS-COV-2-induced cytokine storm [10]. Indeed, the presence of elevated levels of CXCL10 were strongly related with the infiltration of immune cells into alveolar space, peribronchial, and perivascular of the lung [4]. Interestingly, circulating CXCL10 levels were found to be increased in those patients with a more severe clinical course of both COVID-19 as well as of the previous MERS-CoV infection [4].
Yang et al., recently highlighted that CXCL10 levels measured at different time points in COVID-19 patients (i.e. 7 days after diagnosis, between 8 and 14 days and during the recovery phase starting from 15 days after disease onset) were positively correlated with increasing severity (critically ill > severe > moderate) of COVID 19 disease. Furthermore, the circulating levels of CXCL10 were elevated upon admission and remained high during the phase of disease progression. This observation appears of not negligible clinical relevance in that, it would suggest that serum CXCL10 could be also considered a marker for predicting disease progression [32]. In addition, Zhang et al., reported a CXCL10 (and CCL2) positive signature in a significantly higher proportion of macrophages derived from severe COVID-19 than in the other inflamed tissues [33]. More importantly, the Authors, concluded that IFN-γ by activation of CXCL10+ CCL2+ macrophage subset would act as a mediator of severe disease, suggesting that the anti-Type II IFN treatment, including JAK inhibitors, might prove effective for inhibiting the onset of cytokine storm [53,54]. New information were provided also by in a silico study, in which algorithm analysis of the gene set expression based on Core Gene CXCL10 (ssGSEA) showed that CXCL10 is highly expressed in SARS-COV-2 infected patients and that CD4 and CD8 cells are highly enriched in samples with higher expression of CXCL10. On the contrary, samples with low expression of CXCL10 appeared to be enriched in dendritic cells [34]. Moreover Chi et al., showed that, among a panel of chemokines, CXCL10 circulating levels were progressively increased according to a mild, moderate or severe course of COVID-19 [9].
Hue et al., performed an interesting prospective mono-center observational study showing that serum concentrations of CXCL10 and nasopharyngeal viral loads were both positively associated with the outcomes of COVID-19 patients [35]. Indeed, there were dramatically lower serum concentrations of CXCL10 (and some other cytokines and chemokines) in patients having bacterial/non-documented ARDS than in patients with non-COVID-19 viral ARDS or COVID-19, suggesting that CXCL10 and the other chemokines could be biomarkers of viral infections. Moreover, serum concentrations of CXCL10 as well as SARS-COV-2 viral loads remained significantly higher in patients who died than in survivors [35]. Interestingly, they also identified a group of chemokines/cytokines, including CXCL10 that were associated with day-28 mortality in COVID-19 ARDS patients [55]. In a recent retrospective study, evaluating the circulating levels of several chemokine in 74 patients with severe and critical ill COVID-19, CXCL10 and CCL2 were identified as biomarkers positively associated with severity and death-risk of COVID-19. In particular, the circulating concentrations of both CXCL10 and CCL2 in critically ill patients were higher than those found in severe patients. No difference was found in the level of another chemokine (i.e CCL3). Interestingly, stratification of patients according to the d-dimer levels, revealed that the levels of both CXCL10 and CCL2 were higher in patients showing highest d-dimer levels, suggesting that CXCL10 and CCL2 would be related to the risk of death in COVID-19 patients [36].
Blot et al., in BALF from COVID-19 patients, found that, higher CXCL10 levels in both the systemic and alveolar compartments in those patients developing acute respiratory distress syndrome (ARDS) were associated with a longer duration of mechanical ventilation. This finding emphasizes the involvement of the CXCL10-CXCR3 signaling axis in the pathogenesis of severe forms of COVID-19 infection, thus representing a potential therapeutic target [37]. It seems worth remembering that, corticosteroids that showed beneficial effects in COVID-19 patients actually act, through inhibition of the Th1 pathway, upstream of CXCL10. The fact that corticosteroids may reduce CXCL10 secretion was already known [31 [38]. More specifically, Lev et al. reported that a significant decrease of CXCL10 levels was observed in COVID- 19 patients receiving corticosteroids drugs [39]. If further validated, this finding might suggest potentially promising therapeutic approach by specifically neutralizing CXCL10.
5. Chemokines belonging to CC family
5.1. CCL2/ MCP-1 (Monocyte chemoattractant protein-1)
CCL2 is the first discovered human chemokine belonging to the CC chemokine family [40]. CCL2 regulates immune cell recruitment to specific sites. CCL2 preferentially binds the CCR2 receptor, which is expressed in various tissues including blood, brain, heart, kidney, liver, lung, ovary, pancreas, spinal cord, spleen and thymus [41]. CCL2 is constitutively expressed for homeostatic functions such as regulating lymphocyte trafficking from blood to lymph nodes, while it is induced during inflammatory responses when leukocytes are required for tissue defense and repair [42,43]. CCL2 is one of the key chemokines that regulate migration and infiltration of monocytes/macrophages. Several reports demonstrated a dysregulation of CCL2 expression in SARS-COV-2 infected patients. Just to give a few examples, Xiong et al., performed a transcriptome sequencing of the RNAs isolated from the bronchoalveolar lavage fluid and peripheral blood mononuclear cells specimens of COVID-19 patients, reporting an overexpression of CCL2-RNA (in addition to other chemokines-RNAs) [5]. Other studies, confirmed an over-expression of CCL2 in severe COVID-19 patients as compared to the non-severe cases. In particular, Li at al., analyzed blood samples from severe and non-severe COVID-19 patients demonstrating that, significant differences in cell counts of lymphocytes, neutrophils, eosinophils were associated with a profoundly different chemokine profiles between the two groups. Moreover, the circulating levels of CCL2 were nearly doubled (264 vs. 134 pg/mL) in the severe group compared to non-severe group [21]. Also Huang et al., confirmed that intensive care unit (ICU) patients showed significantly higher plasma levels of GSCF, CXCL10, CCL2, CCL3, and TNF-α than those not requiring ICU, further indicating that the occurrence of the “cytokine storms” largely contributes in aggravating the disease, being also associated with the progression of pneumonia/respiratory failure [22]. As previously reported, CCL2 (together with CXCL10) was appear to be associated to disease severity in COVID-19 patients [36]. Higher CCL2 levels were also independently associated (together with other immune soluble biomarkers) with mortality in COVID-19 patients [44]. Kwon et al, confirmed that plasma concentrations of CCL2 (as well as of CXCL10, CXCL8 and CXCL9) were higher in patients with, severe or critical disease, compared with patients with mild disease but, only CCL2, among the cytokines/chemokines analyzed, was positively correlated with the viral load [45]. Finally, Sierra et al., isolated RNA from nasopharyngeal swab samples from COVID-19 positive patients, stratified in very symptomatic, complicated, and fatal cases, showing that CCL2 (and CCL3) expressions were significantly higher in patients with unfavorable outcome compared with cases with a favorable evolution or with negative controls. Moreover, CCL2 higher expression was associated with marked asthenia and with the presence of dyspnea in the unfavorable evolution group. More interestingly the “multivariate beta regression analysis showed that CCL2 was associated with a more severe outcome [46].
5.2. CCL3/ MIP-1-α (macrophage inflammatory protein 1-α)
CCL3 (C-C Motif Chemokine Ligand 3) is a chemokine binding the CCR1, CCR4 and CCR5 chemokine receptors. This chemokine is one of the major HIV-suppressive factors produced by CD8+ T-cells [47]. Recombinant CCL3 was demonstrated to induce a dose-dependent inhibition of different strains of HIV-1, HIV-2, and simian immunodeficiency virus (SIV). Interestingly, it was previously demonstrated that polymorphisms at CCL3 locus are associated with resistance or susceptibility to infection by human immunodeficiency virus type 1 [48].
CCL3 should be included to the list of those chemokines found to be elevated in patients with a more severe prognosis of COVID-9 [5,7,8,49]. It was reported that high levels of some chemokines and of CCL3 in particular, in view of their ability to recruit neutrophils and monocytes, characterize those children developing MISC as a consequence of SARS-COV-2 infection. This finding also suggests that autoreactivity secondary to SARS-COV-2 infection and the inflammatory innate immune response may be critical in the pathogenesis of MIS-C [50]. CCL3 together with other chemokines was found to be highly expressed in PBMC and BALF of COVID-19 patients compared with controls [5]. The increased transcription of CCL2 and CCL3 chemokines receptors CCR2 (i.e. the CCL2 receptor) and CCR5 (i.e. the CCL3 receptor) observed would confirm the activation of the inflammatory signalling promoted by these chemokine/chemokine receptor binding (32). In addition, in the previously cited study by Sierra et al, CCL3 RNA expression level was higher in COVID-19 patients with an unfavourable outcome compared with cases with a favourable evolution or with negative controls and was also associated with asthenia [46]. Of note, Chua et al., investigated the immune response in patients with COVID-19 by single-cell RNA sequencing (scRNA-seq) of nasopharyngeal and bronchial samples, with the aim of identifying specific chemokines/chemokine-receptors genes expression correlating with disease severity. They found a significant induction of CCL2 and CCL3 genes expression in macrophages together with an increased expression of the CCR1 (receptor for both chemokines) gene, in patients with critical COVID-19. The Authors concluded that targeting these chemokine/chemokine receptor binding might suppress immune hyper activation in critical COVID-19 patients [8].
5.3. CCL5 /RANTES (Regulated upon activation, normal t cell expressed and presumably secreted)
CCL5 is a chemokine of the CC chemokine family that binds to CCR1, CCR3, and mainly CCR5 receptors [51]. CCL5 plays an active role in recruiting a variety of leukocytes including T cells, macrophages, eosinophils, and basophils into inflammatory sites. In collaboration with cytokines such as IL-2 and IFN-γ that are released by T cells, CCL5 also induces the activation and proliferation of natural killer cells to generate CC chemokine-activated killer cells [52]. Moreover, it seems that the CCL5/CCR5 binding may favor tumor development by acting as growth factor, stimulating angiogenesis, modulating the extracellular matrix, inducing the recruitment of additional stromal and inflammatory cells, and taking part in tumor immune evasion [53]., It was previously reported that, an increased level of CCL5 (together with other chemokines), could be observed after 24 and 48 h, in mice infected with the previous SARS-CoV [54]. Similarly, the circulating levels of CCL5 seem to play a role also in SARS-COV-2 infection, even if controversial data regarding CCL5 in SARS-COV-2 disease progression are present. Indeed, Li et al., analyzed a panel of blood cytokines in 69 patients with COVID-19, reporting a slight but significant increased CCL5 level, in patients with severe as compared to non-severe COVID-19 [21]. Hue et al., observed a positive correlation between higher CCL5 levels and severe COVID 19 manifestation, highlighting the important role of this chemokine in CS induced ARDS [35]. At difference, Zhao et al. evaluated 71 COVID 19 patients, reporting that circulating CCL5 levels were significantly elevated only in mild cases (in whom they remained elevated for 3/4 week after recovery) as compared with healthy controls. Of note, no elevation of CCL5 circulating levels was observed in the severe group during disease progression, which prompted the Authors to suggest that CCL5 might play a protective role by preventing the development of severe illness in COVID-19 patients [55]. Interestingly enough, Takahashi et al., reported that, among a panel of 71 cytokines/chemokines, CCL5 circulating levels (together with CXCL8 and IL-18 levels) were higher in male compared to female patients affected by SARS-COV-2, suggesting the existence of a gender-dependent effect. Of note some clinical data suggest that treatment with the CCR5 blocking antibody (i.e. Leronlimab) could represent a promising immunotherapy strategy for COVID 19 treatment [56]. More specifically, Patterson et al., reported that treatment with Leronlimab lead to a complete CCR5 receptor occupancy on macrophage and T cells, rapid reduction of plasma IL-6, restoration of the CD4/CD8 ratio, and a significant decrease in SARS-COV-2 plasma viremia [57].
Taking together the above evidences, the role of CCL5 in SARS-COV-2 disease definitely requires corroboration, but further advances in the comprehension of the role of CCL5 could be crucial for better understanding the immunological profile as well as the gender-related differences in the clinical course of SARS-COV-2.
5.4. CCL19/MIP-3-β (macrophage inflammatory protein-3-β)
CCL19 is a pro-inflammatory chemokine that belongs to the CC chemokine family. The binding of CCL19 to its receptor CCR7, mediates several migratory events in adaptive immunity following antigen encounter by immunocytes [58].The binding of CCL19 to CCR7 receptor is crucial for the recruitment of antigen presenting dendritic cells and naïve T-cells to secondary lymphoid organs [59]. CCL19 also plays an important role in rheumatic and autoimmune diseases being overexpressed in autoimmune responses and in chronic inflammatory conditions [58]. Regarding the SARS-COV-2 infection scenario, there are limited data about the role of CCL19, of note Balnis et al. found that high levels of CCL19 chemokine in patients’ serum significantly correlated with outcome and greater mortality in a cohort of patients on mechanical ventilation infected by SASR-COV2 developing ARDS. Based on this findings, the Authors suggested that CCL19 determination could facilitate early identification COVID19-ARDS patients at higher risk of death [60].
5.5. CCL20/MIP-3-α (macrophage inflammatory Protein-3)
CCL20 belongs to the subfamily of CC chemokines and binds to C-C chemokine receptor CCR6. The binding of CCL20 to CCR6 is responsible for the chemotaxis of dendritic cells (DC), lymphocytes, effector/memory T-cells and B-cells and plays an important role at skin and mucosal surfaces in both homeostatic and inflammatory conditions [61]. CCL20 is also involved in cancer inflammation and autoimmune diseases [[61], [62], [63]]. CCL20 is involved in the recruitment of both the pro-inflammatory IL-17 producing helper T-cells (Th17) and the regulatory T-cells (Treg) to the sites of inflammation [61]. CCL20 together with other chemokines, was shown to be involved, in the pathogenesis of COVID-19, being expressed by immune activated macrophages in critical cases of SARS-COV2 infection and being found at higher levels in those patients requiring mechanic ventilation [8]. In addition, CCL20 (similarly to the above described CCL3) was found to be elevated in children affected by MISC as a consequence of SARS-COV-2 infection [50]. Hue et al., reported that CCL20 serum levels (together with other chemokines described in previous paragraphs), were higher in patients with viral ARDS and COVID-19 as compared to non-COVID-19 ARDS. Moreover, increased circulating concentrations of CCL20 (and other chemokines) was registered over time, consistent with the prolonged durations of viral shedding in COVID-19 patients [35].
5.6. The chemokine “immune signature” of patients with COVID-19
The efforts made by the scientific community in recent months have made it clear that COVID-19 displays a kind of “inflammatory signature”, characterized by increased levels of soluble biomarkers (cytokine and chemokines) which are involved in the recruitment and activation of several immune cell types, like monocyte/macrophages, neutrophils, T-lymphocytes, and many others. These immune-active biomarkers, measured either early upon patient admission or throughout hospitalization, may provide clinically relevant information in predicting a more or less severe course of the disease as well as in estimating the mortality risk of infected patients.
Indeed, chemokine dysregulation is a core component of SARS-COV-2 infection, interfering with normal immune-cell chemotaxis and contributing to the recruitment of an excessive and not balanced immune cell milieu in which the virus thrive [64].
The results of the studies discussed in the previews sections highlight how each chemokine (among those evaluated so far) could play a role in COVID-19, not only by driving the cytokine storm but also as immune biomarkers useful for predicting the patient’s outcome.
A major challenge in providing a unifying proposal for a chemokine immune signature in patients with COVID-19 stems from the fact that the currently available studies are characterized by a great heterogeneity in terms of several aspects which include: number of patients enrolled, biological matrix on which chemokines were measured (serum, plasma, BALF, RNA from different types cells, nasopharyngeal samples), the time of measurement in terms of onset of the disease and most importantly which chemokine or chemokine profile pattern was evaluated. The above aspects all contribute in making difficult, if at all possible, direct comparisons of the results provided by each study addressing the issue of which chemokine/chemokine profile better depicts the chemokine signature of patients with COVID-19, as well as the clinical outcome. Thus, drawing a conclusive collective scenario appears not free of the risk of not taking into account that a given study might have identified a given chemokine as the most relevant one just because another chemokine was simply not evaluated.
However, despite the above stated considerations, based on the currently available data, we will attempt to indicate those chemokines that, at least at present, were more firmly identified to represent the “chemokine immune signature” in COVID-19.
By "chemokine immune signature" we are referring to a sort of identity card of a specific circulating chemokine profile of a patient with COVID-19, which would confer a given clinical phenotype of the infection. In particular, the "chemokine immune signature" may hold diagnostic, prognostic and therapeutic implications of patients infected with SARS-COV-2. This could represent an additional tool, helping clinicians in predicting the disease severity and outcome of a given patients and clinical decision making. Based on the currently available studies which were overviewed in the present review, CXCL10 and CCL2 could be reasonably regarded as the two chemokines, for which more solid and consistent data would support a crucial role in determining the clinical course and ultimately the outcome of COVID-19. It should be added that, both CXCL10/CXCR3 and CCL2/CCR2 axes would deserve to be more deeply investigated in terms of their potential as therapeutic targets.
The idea that CXCL10 and CCL2 should be regarded as “dominant” in the chemokine immune signature stems from the number of studies consistently reporting their crucial role in the onset of the cytokine storm of COVID-19 and from their relevance in determining specific features of this condition. Indeed, among the several chemokines which have been evaluated and found to be involved in COVID-19, CXCL10 and CCL2 were found to be the most up-regulated ones as compared to controls, showing different degrees of correlation with the viral load as well as with several peculiar symptoms (asthenia, dyspnea, anosmia) [5,[7], [8], [9], [10], [11],[21], [22], [23],29,36,37,[44], [45], [46],49,54,[65], [66], [67]]. Most importantly, CXCL10 and CCL2 were identified to be rather good markers of disease severity since their circulating levels progressively increase in patients with moderate, mild and severe clinical manifestations. Furthermore, CXCL10 and CCL2 were considered potential initiators of the deadly COVID-19 immuno-pathological pathway being correlated to increased risk of mortality [11,13]. From a biological point of view, the circulating levels of these chemokines crucially determine the recruitment of a specific immune cell milieu infiltrating lungs in COVID-19 patients. As previously stated, the type and the abundance of cell infiltrate will be crucial in the development of the feedback loop characterizing the cytokine storm. In particular, on one hand, CXCL10 recruits monocytes and macrophages expressing CXCR3 sustaining the early Th1 oriented immune response of COVID-19 patients [66], on the other hand, CXCL10 by selectively binding endothelial cells expressing the CXCR3 receptor, will impair endothelial integrity [68], which is now known to be a major determinant of COVID-19 severe clinical consequences [69]. At the same time, CCL2 binds the CCR2 and CCR4 receptors that are expressed on monocytes, macrophages and basophiles being ultimately recruited by this chemokine. The elevated levels of CCL2 found in post mortem autopsy are in line with the abundant presence of peripherally derived monocytes and macrophages in the lungs of severe cases of COVID-19 [7] which are now known to play an important role in driving resident cell damage [70].
From a clinical point of view, the notion that chemokine levels in the early phase of the infection seem reliable predictors of patient outcome, raise the relevant issue of using serum chemokine levels for therapeutic decision-making. The predictive value of chemokines for early identification of patients more prone to experience respiratory failure, organ damage and eventually death, might prove helpful for choosing different therapeutic interventions as well as to design future clinical trials aimed at limiting the underlying inflammatory milieu. Furthermore, a prediction model based on chemokine levels early in disease might also provide relevant information for healthcare in allocation and prioritization of the highest-risk patients. As a final consideration, given that it seems reasonable to propose that early immunological interventions targeting inflammatory markers specifically predicting a more severe disease outcome, could be more effective than those modulating late-appearing chemokines.
6. Conclusions
In the last few months, rapidly growing evidences supporting an important role of chemokines in the onset, progression and recovery of SARS-COV-2 infection have accumulated. Since our last review article on the topic of chemokine involvement in COVID-19-related cytokine storm, it has become clear now, that specific chemokines are involved at given times of the infection and that changes in their levels may reflect different clinical course of SARS-COV-2 infection. Hopefully, the knowledge derived from these studies may help identifying those patients carrying higher risk to develop severe COVID-19 and be useful for the identification of biomarkers for individual risk stratification. The development and optimization of therapeutic strategies aimed at targeting chemokines and/or their receptors could add relevant tools for the clinical management of patients with COVID-19. Several chemokines and their relation with SARS-COV-2 infection have been evaluated so far. In the present review article, an overview of the current findings for each chemokine is provided. At present, CXCL10 among the CXC family and CCL2 among the CC family appear to be the most extensively studied ones, as well as those with particular clinical relevance in view of the strong correlation between their circulating concentrations and the severity of SARS-COV-2 infection.
A more precise characterization of the chemokine immune signature will hopefully provide more insights into the role of early measurable markers for predicting disease progression and length of hospitalization as well as for choosing targeted therapies. The possibility to build up a combined routine clinical tests, simultaneously evaluating one or more circulating chemokine (and/or other soluble markers), could be envisaged to improve early risk-based stratification of patients, with relevant benefit especially when healthcare systems become over-stretched.
In conclusion, advances in our understanding the physiopathology of SARS-COV-2 infection strongly support the crucial role of chemokines in COVID-19. In the last few months, a rapidly growing number of evidences have accumulated and likely will increase in short time. Future studies might confirm currently available results and/or provide further insights on the role of other chemokines playing an important role in COVID-19. The present review does not pretend to be conclusive as to the complex mechanisms sustaining the cytokine storm in COVID-19, nevertheless, thanks to the efforts of the Scientific Community, we might be at a good starting point for a more precise characterization of the "immune signature" of COVID-19, hopefully being not so far from the finish line.
Funding
This paper was not supported by any grant or funding.
Declarations of Competing Interest
None.
Biographies

Francesca Coperchini completed her University education in 2009 obtaining the degree of Specialized Biologist at University of Pavia. She is Specialist in Clinical Pathology at University of Pavia and she is Researcher Biologist at Laboratory for Endocrine Disruptors, ICS Maugeri SpA –I.R.C.C.S, Pavia. Main Research fields: Role of chemokines in autoimmune endocrine disorders and thyroid cancer, endocrine disruptors.

Luca Chiovato obtained his M.D. degree at the University of Pisa in 1976. He completed his postgraduate Degree in Internal Medicine in 1982 and obtained his PhD in Endocrine and Metabolic Sciences at University of Florence in 1988. Present position: Full Professor of Endocrinology at University of Pavia, Director of the Internal Medicine and Therapeutics Department of the University of Pavia, Director of the Laboratory for Endocrine Disruptors and Director of the Head Unit of Internal Medicine and Endocrinology, ICS Maugeri SpA –I.R.C.C.S, Pavia. Main research fields: endocrine autoimmunity; thyroid diseases, genetics and clinics of obesity; endocrine disruptors.

Gianluca Ricci obtained the degree of Specialized Biologist at University of Pavia in 2019. He is Researcher Biologist at Laboratory for Endocrine Disruptors, ICS Maugeri SpA –I.R.C.C.S, Pavia.

Laura Croce obtained her M.D. degree at the University of Pavia in 2013. She completed her postgraduate Degree in Endocrinology and Metabolism in 2018 at University of Pavia. Present Position: Researcher in Endocrinology and Metabolism at the University of Pavia. Main Research fields: clinics of thyroid disease, role of chemokines in autoimmune endocrine disorders and thyroid cancer, endocrine disruptors, obesity.

Flavia Magri obtained his M.D. degree at the University of Pavia in 1987. She completed her postgraduate Degree in Endocrinology in 1990 and in Internal Medicine in 1995. She obtained her PhD in Internal Medicine and Medical Therapy at University of Pavia in 2000. Present position: Associate Professor in Endocrinology at University of Pavia. She works at the Unit of Internal Medicine and Endocrinology, ICS Maugeri SpA – I.R.C.C.S, Pavia. Main research fields are: clinics of thyroid diseases, thyroid cancer and thyroid diseases in pregnancy

Mario Rotondi obtained his M.D. degree at the University of Naples in 1992. He completed his postgraduate Degree in Endocrinology and Metabolism at the University of Naples in 1998 and obtained his PhD in Endocrine and Metabolic Sciences at University of Naples in 2003 with a project focused on the role of chemokines in Graves’ disease. Present position: Associate Professor in Endocrinology at University of Pavia, Director of the School of Postgraduate Endocrinology and Metabolism, University of Pavia. He works at the Laboratory for Endocrine Disruptors and at the Unit of Internal Medicine and Endocrinology, ICS Maugeri SpA – I.R.C.C.S, Pavia. Main research fields are: physiopathology and clinics of thyroid diseases, thyroid autoimmunity; thyroid diseases in pregnancy; role of chemokines in autoimmune endocrine disorders, thyroid cancer.
References
- 1.W.H. Organization . 2020. Coronavirus Disease (COVID-19) Pandemic.https://www.who.int/emergencies/diseases/novel-coronavirus-2019 [Google Scholar]
- 2.Kaiko G.E., Horvat J.C., Beagley K.W., Hansbro P.M. Immunological decision-making: how does the immune system decide to mount a helper T-cell response? Immunology. 2008;123(3):326–338. doi: 10.1111/j.1365-2567.2007.02719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sokol C.L., Luster A.D. The chemokine system in innate immunity. Cold Spring Harb. Perspect. Biol. 2015;7(5) doi: 10.1101/cshperspect.a016303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., Wu H., Lin Y., Zhang M., Zhang Q., Shi M., Zhou Y., Lan K., Chen Y. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., Yang D., Zhang G., Li H., Chen F., Xu Y., Chen M., Gao Z., Dong J., Liu B., Zhang X., Wang W., He K., Jin Q., Li M., Wang J. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27(6) doi: 10.1016/j.chom.2020.04.017. 883-890.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., Liu L., Amit I., Zhang S., Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 7.Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thürmann L., Kurth F., Völker M.T., Kazmierski J., Timmermann B., Twardziok S., Schneider S., Machleidt F., Müller-Redetzky H., Maier M., Krannich A., Schmidt S., Balzer F., Liebig J., Loske J., Suttorp N., Eils J., Ishaque N., Liebert U.G., von Kalle C., Hocke A., Witzenrath M., Goffinet C., Drosten C., Laudi S., Lehmann I., Conrad C., Sander L.E., Eils R. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38(8):970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 8.Chi Y., Ge Y., Wu B., Zhang W., Wu T., Wen T., Liu J., Guo X., Huang C., Jiao Y., Zhu F., Zhu B., Cui L. Serum Cytokine and Chemokine Profile in Relation to the Severity of Coronavirus Disease 2019 in China. J. Infect. Dis. 2020;222(5):746–754. doi: 10.1093/infdis/jiaa363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., Takahashi T., Tokuyama M., Lu P., Venkataraman A., Park A., Mohanty S., Wang H., Wyllie A.L., Vogels C.B.F., Earnest R., Lapidus S., Ott I.M., Moore A.J., Muenker M.C., Fournier J.B., Campbell M., Odio C.D., Casanovas-Massana A., Herbst R., Shaw A.C., Medzhitov R., Schulz W.L., Grubaugh N.D., Dela Cruz C., Farhadian S., Ko A.I., Omer S.B., Iwasaki A., Team Y.I. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A. B.R. tenOever, imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181(5) doi: 10.1016/j.cell.2020.04.026. 1036-1045.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffré F., Zhang T., Kim T.W., Harschnitz O., Redmond D., Houghton S., Liu C., Naji A., Ciceri G., Guttikonda S., Bram Y., Nguyen D.T., Cioffi M., Chandar V., Hoagland D.A., Huang Y., Xiang J., Wang H., Lyden D., Borczuk A., Chen H.J., Studer L., Pan F.C., Ho D.D. B.R. tenOever, T. Evans, R.E. Schwartz, S. Chen, A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27(1) doi: 10.1016/j.stem.2020.06.015. 125-136.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu L., Zhang H., Zhan M., Jiang J., Yin H., Dauphars D.J., Li S.Y., Li Y., He Y.W. Preventing Mortality in COVID-19 Patients: Which Cytokine to Target in a Raging Storm? Front. Cell Dev. Biol. 2020;8:677. doi: 10.3389/fcell.2020.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogata H., Sekikawa A., Yamagishi H., Ichikawa K., Tomita S., Imura J., Ito Y., Fujita M., Tsubaki M., Kato H., Fujimori T., Fukui H. GROα promotes invasion of colorectal cancer cells. Oncol. Rep. 2010;24(6):1479–1486. [PubMed] [Google Scholar]
- 14.Anisowicz A., Bardwell L., Sager R. Constitutive overexpression of a growth-regulated gene in transformed Chinese hamster and human cells. Proc Natl Acad Sci U S A. 1987;84(20):7188–7192. doi: 10.1073/pnas.84.20.7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghebremedhin A., Salam A.B., Adu-Addai B., Noonan S., Stratton R., Ahmed M.S.U., Khantwal C., Martin G.R., Lin H., Andrews C., Karanam B., Rudloff U., Lopez H., Jaynes J., Yates C. A novel CD206 targeting peptide inhibits bleomycin induced pulmonary fibrosis in mice. bioRxiv. 2020 doi: 10.3390/cells12091254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rotondi M., Coperchini F., Chiovato L. CXCL8 in thyroid disease: from basic notions to potential applications in clinical practice. Cytokine Growth Factor Rev. 2013;24(6):539–546. doi: 10.1016/j.cytogfr.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Rotondi M., Coperchini F., Latrofa F., Chiovato L. Role of Chemokines in Thyroid Cancer Microenvironment: Is CXCL8 the Main Player? Front. Endocrinol. (Lausanne) 2018;9:314. doi: 10.3389/fendo.2018.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanderheiden A., Ralfs P., Chirkova T., Upadhyay A.A., Zimmerman M.G., Bedoya S., Aoued H., Tharp G.M., Pellegrini K.L., Manfredi C., Sorscher E., Mainou B., Lobby J.L., Kohlmeier J.E., Lowen A.C., Shi P.Y., Menachery V.D., Anderson L.J., Grakoui A., Bosinger S.E., Suthar M.S. Type I and type III interferons restrict SARS-CoV-2 infection of human airway epithelial cultures. J. Virol. 2020;94(19) doi: 10.1128/JVI.00985-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y., Liao W., Wan L., Xiang T., Zhang W. Correlation between relative nasopharyngeal virus RNA load and lymphocyte count disease severity in patients with COVID-19. Viral Immunol. 2020 doi: 10.1089/vim.2020.0062. [DOI] [PubMed] [Google Scholar]
- 20.Li S., Jiang L., Li X., Lin F., Wang Y., Li B., Jiang T., An W., Liu S., Liu H., Xu P., Zhao L., Zhang L., Mu J., Wang H., Kang J., Li Y., Huang L., Zhu C., Zhao S., Lu J., Ji J., Zhao J. Clinical and pathological investigation of patients with severe COVID-19. JCI Insight. 2020;5(12) doi: 10.1172/jci.insight.138070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tincati C., Cannizzo E.S., Giacomelli M., Badolato R., d’Arminio Monforte A., Marchetti G. Heightened circulating interferon-inducible chemokines, and activated pro-cytolytic Th1-Cell phenotype features Covid-19 aggravation in the second week of illness. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.580987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diorio C., Henrickson S.E., Vella L.A., McNerney K.O., Chase J., Burudpakdee C., Lee J.H., Jasen C., Balamuth F., Barrett D.M., Banwell B.L., Bernt K.M., Blatz A.M., Chiotos K., Fisher B.T., Fitzgerald J.C., Gerber J.S., Gollomp K., Gray C., Grupp S.A., Harris R.M., Kilbaugh T.J., John A.R.O., Lambert M., Liebling E.J., Paessler M.E., Petrosa W., Phillips C., Reilly A.F., Romberg N.D., Seif A., Sesok-Pizzini D.A., Sullivan K.E., Vardaro J., Behrens E.M., Teachey D.T., Bassiri H. Multisystem inflammatory syndrome in children and COVID-19 are distinct presentations of SARS-CoV-2. J. Clin. Invest. 2020 doi: 10.1172/JCI140970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Del Valle D.M., Kim-Schulze S., Hsin-Hui H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T., Madduri D., Stock A., Marron T., Xie H., Patel M.K., van Oekelen O., Rahman A., Kovatch P., Aberg J., Schadt E., Jagannath S., Mazumdar M., Charney A., Firpo-Betancourt A., Mendu D.R., Jhang J., Reich D., Sigel K., Cordon-Cardo C., Feldmann M., Parekh S., Merad M., Gnjatic S. An inflammatory cytokine signature helps predict COVID-19 severity and death. medRxiv. 2020 doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bilusic M., Heery C.R., Collins J.M., Donahue R.N., Palena C., Madan R.A., Karzai F., Marté J.L., Strauss J., Gatti-Mays M.E., Schlom J., Gulley J.L. Phase I trial of HuMax-IL8 (BMS-986253), an anti-IL-8 monoclonal antibody, in patients with metastatic or unresectable solid tumors. J. Immunother. Cancer. 2019;7(1):240. doi: 10.1186/s40425-019-0706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tokunaga R., Zhang W., Naseem M., Puccini A., Berger M.D., Soni S., McSkane M., Baba H., Lenz H.J. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation - A target for novel cancer therapy. Cancer Treat. Rev. 2018;63:40–47. doi: 10.1016/j.ctrv.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rotondi M., Chiovato L., Romagnani S., Serio M., Romagnani P. Role of chemokines in endocrine autoimmune diseases. Endocr. Rev. 2007;28(5):492–520. doi: 10.1210/er.2006-0044. [DOI] [PubMed] [Google Scholar]
- 28.Moratto D., Giacomelli M., Chiarini M., Savarè L., Saccani B., Motta M., Timpano S., Poli P., Paghera S., Imberti L., Cannizzo S., Quiros-Roldan E., Marchetti G., Badolato R. Immune response in children with COVID-19 is characterized by lower levels of T-cell activation than infected adults. Eur. J. Immunol. 2020 doi: 10.1002/eji.202048724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belperio J.A., Keane M.P., Arenberg D.A., Addison C.L., Ehlert J.E., Burdick M.D., Strieter R.M. CXC chemokines in angiogenesis. J. Leukoc. Biol. 2000;68(1):1–8. [PubMed] [Google Scholar]
- 30.Romagnani P., Maggi L., Mazzinghi B., Cosmi L., Lasagni L., Liotta F., Lazzeri E., Angeli R., Rotondi M., Filì L., Parronchi P., Serio M., Maggi E., Romagnani S., Annunziato F. CXCR3-mediated opposite effects of CXCL10 and CXCL4 on TH1 or TH2 cytokine production. J. Allergy Clin. Immunol. 2005;116(6) doi: 10.1016/j.jaci.2005.09.035. 1372-9. [DOI] [PubMed] [Google Scholar]
- 31.Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020 doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y., Shen C., Li J., Yuan J., Wei J., Huang F., Wang F., Li G., Li Y., Xing L., Peng L., Yang M., Cao M., Zheng H., Wu W., Zou R., Li D., Xu Z., Wang H., Zhang M., Zhang Z., Gao G.F., Jiang C., Liu L., Liu Y. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J. Allergy Clin. Immunol. 2020 doi: 10.1016/j.jaci.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang F., Mears J.R., Shakib L., Beynor J.I., Shanaj S., Korsunsky I., Nathan A., Donlin L.T., Raychaudhuri S. Ifn- bioRxiv. 2020 [Google Scholar]
- 34.Zhang N., Zhao Y.D., Wang X.M. CXCL10 an important chemokine associated with cytokine storm in COVID-19 infected patients. Eur. Rev. Med. Pharmacol. Sci. 2020;24(13):7497–7505. doi: 10.26355/eurrev_202007_21922. [DOI] [PubMed] [Google Scholar]
- 35.Hue S., Beldi-Ferchiou A., Bendib I., Surenaud M., Fourati S., Frapard T., Rivoal S., Razazi K., Carteaux G., Delfau-Larue M.H., Mekontso Dessap A., Audureau E., de Prost N. Uncontrolled innate and impaired adaptive immune responses in patients with COVID-19 ARDS. Am. J. Respir. Crit. Care Med. 2020 doi: 10.1164/rccm.202005-1885OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y., Wang J., Liu C., Su L., Zhang D., Fan J., Yang Y., Xiao M., Xie J., Xu Y., Li Y., Zhang S. IP-10 and MCP-1 as biomarkers associated with disease severity of COVID-19. Mol Med. 2020;26(1):97. doi: 10.1186/s10020-020-00230-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blot M., Jacquier M., Aho Glele L.S., Beltramo G., Nguyen M., Bonniaud P., Prin S., Andreu P., Bouhemad B., Bour J.B., Binquet C., Piroth L., Pais de Barros J.P., Masson D., Quenot J.P., Charles P.E., P.s. group CXCL10 could drive longer duration of mechanical ventilation during COVID-19 ARDS. Crit Care. 2020;24(1):632. doi: 10.1186/s13054-020-03328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rotondi M., Falorni A., De Bellis A., Laureti S., Ferruzzi P., Romagnani P., Buonamano A., Lazzeri E., Crescioli C., Mannelli M., Santeusanio F., Bellastella A., Serio M. Elevated serum interferon-gamma-inducible chemokine-10/CXC chemokine ligand-10 in autoimmune primary adrenal insufficiency and in vitro expression in human adrenal cells primary cultures after stimulation with proinflammatory cytokines. J. Clin. Endocrinol. Metab. 2005;90(4):2357–2363. doi: 10.1210/jc.2004-1062. [DOI] [PubMed] [Google Scholar]
- 39.Lev S. Real-time IP-10 measurements as a new tool for inflammation regulation within a clinical decision support protocol for managing severe COVID-19 patients, in: T. Gottesman, G.S. Levin, D. Lederfein, E. Berkov, D. Diker, A. Zaidman, e.a. A. Nutman (Eds.) medRxiv preprint. 2020 [Google Scholar]
- 40.Van Coillie E., Van Damme J., Opdenakker G. The MCP/eotaxin subfamily of CC chemokines. Cytokine Growth Factor Rev. 1999;10(1):61–86. doi: 10.1016/s1359-6101(99)00005-2. [DOI] [PubMed] [Google Scholar]
- 41.Lim S.Y., Yuzhalin A.E., Gordon-Weeks A.N., Muschel R.J. Targeting the CCL2-CCR2 signaling axis in cancer metastasis. Oncotarget. 2016;7(19):28697–28710. doi: 10.18632/oncotarget.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rollins B.J. Chemokines. Blood. 1997;90(3):909–928. [PubMed] [Google Scholar]
- 43.Deshmane S.L., Kremlev S., Amini S., Sawaya B.E. Monocyte chemoattractant protein-1 (MCP-1): an overview. J. Interferon Cytokine Res. 2009;29(6):313–326. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abers M.S., Delmonte O.M., Ricotta E.E., Fintzi J., Fink D., de Jesus A.A., Zarember K.A., Alehashemi S., Oikonomou V., Desai J.V., Canna S.W., Shakoory B., Dobbs K., Imberti L., Sottini A., Quiros-Roldan E., Castelli F., Rossi C., Brugnoni D., Biondi A., Bettini L.R., D’Angio’ M., Bonfanti P., Castagnoli R., Montagna D., Licari A., Marseglia G.L., Gliniewicz E., Shaw E.R., Kahle D., Rastegar A.T., Stack M.A., Myint-Hpu K., Levinson S.L., DiNubile M.J., Chertow D.W., Burbelo P., Cohen J.I., Calvo K.R., Tsang J.S., Su H.C., Gallin J.I., Kuhns D.B., Goldbach-Mansky R., Lionakis M.S., Notarangelo L.D. An immune-based biomarker signature is associated with mortality in COVID-19 patients. JCI Insight. 2020 doi: 10.1172/jci.insight.144455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwon J.S., Kim J.Y., Kim M.C., Park S.Y., Kim B.N., Bae S., Cha H.H., Jung J., Kim M.J., Lee M.J., Choi S.H., Chung J.W., Shin E.C., Kim S.H. Factors of severity in patients with COVID-19: Cytokine/Chemokine concentrations, viral load, and antibody responses. Am. J. Trop. Med. Hyg. 2020;103(6):2412–2418. doi: 10.4269/ajtmh.20-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sierra B., Pérez A.B., Aguirre E., Bracho C., Valdés O., Jimenez N., Baldoquin W., Gonzalez G., Ortega L.M., Montalvo M.C., Resik S., Alvarez D., Guzmán M.G. Association of early nasopharyngeal immune markers with COVID-19 clinical outcome: predictive value of CCL2/MCP-1. Open Forum Infect. Dis. 2020;7(10) doi: 10.1093/ofid/ofaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maurer M., von Stebut E. Macrophage inflammatory protein-1. Int. J. Biochem. Cell Biol. 2004;36(10):1882–1886. doi: 10.1016/j.biocel.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 48.Canque B., Rosenzwajg M., Gey A., Tartour E., Fridman W.H., Gluckman J.C. Macrophage inflammatory protein-1alpha is induced by human immunodeficiency virus infection of monocyte-derived macrophages. Blood. 1996;87(5):2011–2019. [PubMed] [Google Scholar]
- 49.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gruber C.N., Patel R.S., Trachtman R., Lepow L., Amanat F., Krammer F., Wilson K.M., Onel K., Geanon D., Tuballes K., Patel M., Mouskas K., O’Donnell T., Merritt E., Simons N.W., Barcessat V., Del Valle D.M., Udondem S., Kang G., Gangadharan S., Ofori-Amanfo G., Laserson U., Rahman A., Kim-Schulze S., Charney A.W., Gnjatic S., Gelb B.D., Merad M., Bogunovic D. Mapping systemic inflammation and antibody responses in multisystem inflammatory syndrome in children (MIS-C) Cell. 2020 doi: 10.1016/j.cell.2020.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aldinucci D., Colombatti A. The inflammatory chemokine CCL5 and cancer progression. Mediators Inflamm. 2014;2014 doi: 10.1155/2014/292376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soria G., Ben-Baruch A. The inflammatory chemokines CCL2 and CCL5 in breast cancer. Cancer Lett. 2008;267(2):271–285. doi: 10.1016/j.canlet.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 53.Kershaw M.H., Westwood J.A., Darcy P.K. Gene-engineered T cells for cancer therapy. Nat. Rev. Cancer. 2013;13(8):525–541. doi: 10.1038/nrc3565. [DOI] [PubMed] [Google Scholar]
- 54.Yao Z., Zheng Z., Wu K., Junhua Z. Immune environment modulation in pneumonia patients caused by coronavirus: SARS-CoV, MERS-CoV and SARS-CoV-2. Aging (Albany NY) 2020;12(9):7639–7651. doi: 10.18632/aging.103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao Y., Qin L., Zhang P., Li K., Liang L., Sun J., Xu B., Dai Y., Li X., Zhang C., Peng Y., Feng Y., Li A., Hu Z., Xiang H., Ogg G., Ho L.P., McMichael A., Jin R., Knight J.C., Dong T., Zhang Y. Longitudinal COVID-19 profiling associates IL-1RA and IL-10 with disease severity and RANTES with mild disease. JCI Insight. 2020;5(13) doi: 10.1172/jci.insight.139834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takahashi T., Ellingson M.K., Wong P., Israelow B., Lucas C., Klein J., Silva J., Mao T., Oh J.E., Tokuyama M., Lu P., Venkataraman A., Park A., Liu F., Meir A., Sun J., Wang E.Y., Casanovas-Massana A., Wyllie A.L., Vogels C.B.F., Earnest R., Lapidus S., Ott I.M., Moore A.J., Shaw A., Fournier J.B., Odio C.D., Farhadian S., Dela Cruz C., Grubaugh N.D., Schulz W.L., Ring A.M., Ko A.I., Omer S.B., Iwasaki A., Team Y.I.R. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020 doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patterson B.K., Seethamraju H., Dhody K., Corley M.J., Kazempour K., Lalezari J.P., Pang A.P., Sugai C., Francisco E.B., Pise A., Rodrigues H., Ryou M., Wu H.L., Webb G.M., Park B.S., Kelly S., Pourhassan N., Lelic A., Kdouh L., Herrera M., Hall E., Aklin E., Ndhlovu L., Sacha J.B. Disruption of the CCL5/RANTES-CCR5 pathway restores immune homeostasis and reduces plasma viral load in critical COVID-19. medRxiv. 2020 [Google Scholar]
- 58.Yan Y., Chen R., Wang X., Hu K., Huang L., Lu M., Hu Q. CCL19 and CCR7 expression, signaling pathways, and adjuvant functions in viral infection and prevention. Front. Cell Dev. Biol. 2019;7:212. doi: 10.3389/fcell.2019.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Veldkamp C.T., Kiermaier E., Gabel-Eissens S.J., Gillitzer M.L., Lippner D.R., DiSilvio F.A., Mueller C.J., Wantuch P.L., Chaffee G.R., Famiglietti M.W., Zgoba D.M., Bailey A.A., Bah Y., Engebretson S.J., Graupner D.R., Lackner E.R., LaRosa V.D., Medeiros T., Olson M.L., Phillips A.J., Pyles H., Richard A.M., Schoeller S.J., Touzeau B., Williams L.G., Sixt M., Peterson F.C. Solution structure of CCL19 and identification of overlapping CCR7 and PSGL-1 binding sites. Biochemistry. 2015;54(27):4163–4166. doi: 10.1021/acs.biochem.5b00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Balnis J., Adam A.P., Chopra A., Chieng H.C., Feustel P.J., Overmyer K.A., Shishkova E., Coon J.C., Singer H.A., Judson M.A., Jaitovich A. Higher plasma levels of Chemokine CCL19 are associated with poor SARS-CoV-2 acute respiratory distress syndrome (ARDS) outcomes. medRxiv. 2020 [Google Scholar]
- 61.Nelson R.T., Boyd J., Gladue R.P., Paradis T., Thomas R., Cunningham A.C., Lira P., Brissette W.H., Hayes L., Hames L.M., Neote K.S., McColl S.R. Genomic organization of the CC chemokine mip-3alpha/CCL20/larc/exodus/SCYA20, showing gene structure, splice variants, and chromosome localization. Genomics. 2001;73(1):28–37. doi: 10.1006/geno.2001.6482. [DOI] [PubMed] [Google Scholar]
- 62.Ito T., Carson W.F., Cavassani K.A., Connett J.M., Kunkel S.L. CCR6 as a mediator of immunity in the lung and gut. Exp. Cell Res. 2011;317(5):613–619. doi: 10.1016/j.yexcr.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hieshima K., Imai T., Opdenakker G., Van Damme J., Kusuda J., Tei H., Sakaki Y., Takatsuki K., Miura R., Yoshie O., Nomiyama H. Molecular cloning of a novel human CC chemokine liver and activation-regulated chemokine (LARC) expressed in liver. Chemotactic activity for lymphocytes and gene localization on chromosome 2. J. Biol. Chem. 1997;272(9):5846–5853. doi: 10.1074/jbc.272.9.5846. [DOI] [PubMed] [Google Scholar]
- 64.Laing A.G., Lorenc A., Del Molino Del Barrio I., Das A., Fish M., Monin L., Muñoz-Ruiz M., McKenzie D.R., Hayday T.S., Francos-Quijorna I., Kamdar S., Joseph M., Davies D., Davis R., Jennings A., Zlatareva I., Vantourout P., Wu Y., Sofra V., Cano F., Greco M., Theodoridis E., Freedman J., Gee S., Chan J.N.E., Ryan S., Bugallo-Blanco E., Peterson P., Kisand K., Haljasmägi L., Chadli L., Moingeon P., Martinez L., Merrick B., Bisnauthsing K., Brooks K., Ibrahim M.A.A., Mason J., Lopez Gomez F., Babalola K., Abdul-Jawad S., Cason J., Mant C., Seow J., Graham C., Doores K.J., Di Rosa F., Edgeworth J., Shankar-Hari M., Hayday A.C. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020 doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 65.Blot M., Bour J.B., Quenot J.P., Bourredjem A., Nguyen M., Guy J., Monier S., Georges M., Large A., Dargent A., Guilhem A., Mouries-Martin S., Barben J., Bouhemad B., Charles P.E., Chavanet P., Binquet C., Piroth L., L.s. group The dysregulated innate immune response in severe COVID-19 pneumonia that could drive poorer outcome. J. Transl. Med. 2020;18(1):457. doi: 10.1186/s12967-020-02646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bodnar R.J., Yates C.C., Rodgers M.E., Du X., Wells A. IP-10 induces dissociation of newly formed blood vessels. J. Cell. Sci. 2009;122(Pt 12):2064–2077. doi: 10.1242/jcs.048793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang Yang., Shen Chenguang, Li Jinxiu, Yuan Jing, Yang M., F.W., G.L., Y.L, Xing Li, Peng Ling, Wei Jinli, Cao Mengli, Zheng Haixia, Wu Weibo, Zou Rongrong, Li Delin, Xu Zhixiang, Wang Haiyan, Zhang Mingxia, Zhang Zheng, Liu Lei, Liu Y. 2020. Exuberant Elevation of IP-10, MCP-3 and IL-1ra During SARS-CoV-2 Infection Is Associated With Disease Severity and Fatal Outcome. [Google Scholar]
- 68.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20(6):355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]