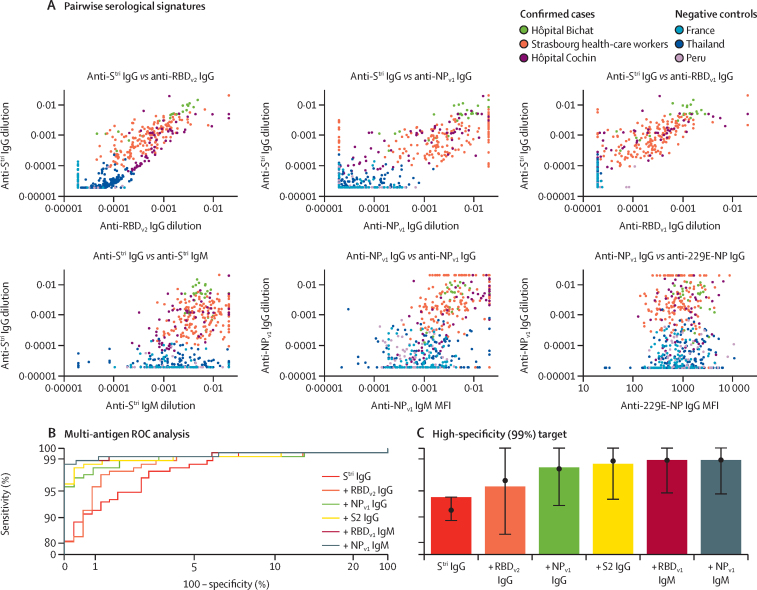
Figure 2.
Serological signatures of SARS-CoV-2 infection
(A) Pairwise combinations of antibody responses. Each point denotes a measured antibody response from a sample from Hôpital Bichat (n=34), health-care workers from Nouvel Hôpital Civil and Hôpital de Haute Pierre in Strasbourg (n=162), and Hôpital Cochin (n=63). Negative control samples are included from Thailand (n=68), Peru (n=90), and French blood donors (n=177). (B) ROC curves for multiple biomarker classifiers generated using a random forests algorithm. Biomarkers are added sequentially. The axes have been rescaled to better differentiate between high values of sensitivity and specificity. (C) For a high specificity target (>99%), sensitivity increases with additional biomarkers, added sequentially. Sensitivity was estimated using a random forests classifier. Points and whiskers denote the median and 95% CIs from repeat cross-validation. MFI= median fluorescent intensity. NP=SARS-CoV-2 nucleoprotein. RBD=SARS-CoV-2 spike glycoprotein receptor-binding domain. ROC=receiver operating characteristic. SARS-CoV-2=severe acute respiratory syndrome coronavirus 2. Stri=SARS-CoV-2 trimeric spike protein. S2=SARS-CoV-2 spike glycoprotein (S2 domain). 229E-NP=human coronavirus 229E NP.