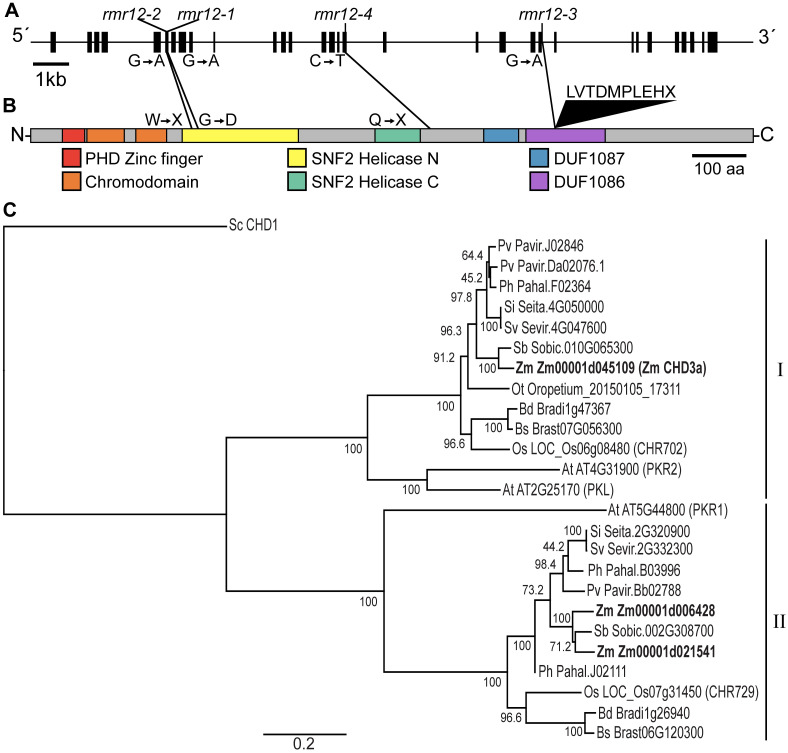
Fig 2. rmr12 mutations disrupt a CHD3-encoding gene model.
(A) Maize gene model Zm00001d045109 highlighting transition mutations found in cDNA sequences from rmr12-1, rmr12-2, rmr12-3, and rmr12-4 mutants. (B) Predicted protein model highlighting domains diagnostic of CHD3 proteins and missense, nonsense (X), and insertion lesions corresponding to the respective transition mutations in (A). (C) Maximum likelihood tree produced from alignment of full-length maize (Zm) CHD3 protein sequences with CHD3 proteins from Arabidopsis (At) and grasses including Brachypodium distachyon (Bd), Brachypodium stacei (Bs), Oropetium thomaeum (Ot), rice (Os), Panicum hallii (Ph), Panicum virgatum (Pv), Setaria italica (Si), Setaria viridis (Sv), and Sorghum bicolor (Sb) identifies two clades (I and II). The tree is anchored with Saccharomyces cerevisiae (Sc) CHD1. Branch lengths depict substitutions per site.