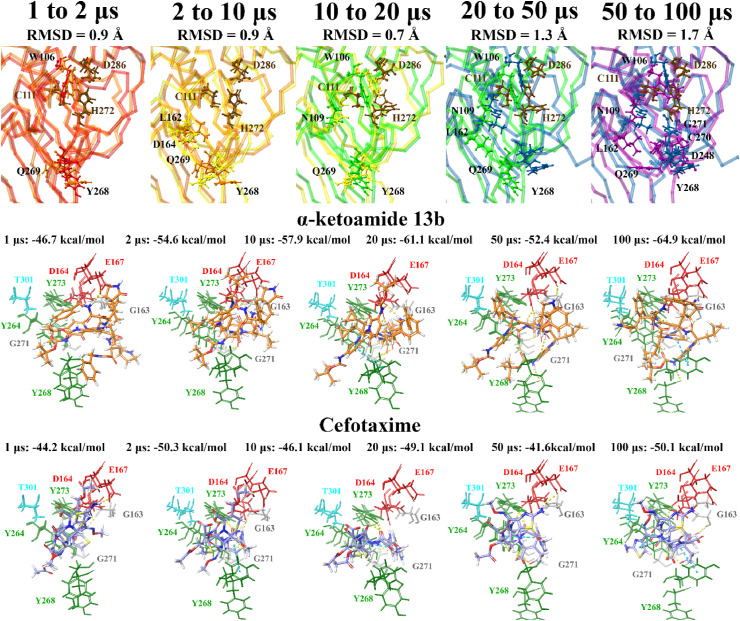
Fig. 5.
Docking of lead pharmacological compounds to snapshots of a 100 μs dynamic simulation of PLpro (100 μs PDB 6WX4; DESRES-ANTON-11441075). Molecular docking was performed to multiple snapshots of a dynamic simulation of PLpro, with comparisons between the different time points shown. Binding site residues were aligned, and RMSDs between time points are stated. Residues with an RMSD greater than 2 following alignment are labelled and highlighted in a ball and stick representation. Lead pharmacological compounds docked to the naphthalene inhibitor binding pocket are depicted, with Glide Energies for docking to each structure stated. Residue interactions are shown as dashed lines, including hydrogen bonds (yellow), salt bridges (magenta), π−π interactions (cyan), and π−cation interactions (green). Amino acid residues are coloured according to their properties, namely hydrophobic residues (green), polar uncharged residues (cyan), negatively charged residues (red), and glycine residues (grey). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)