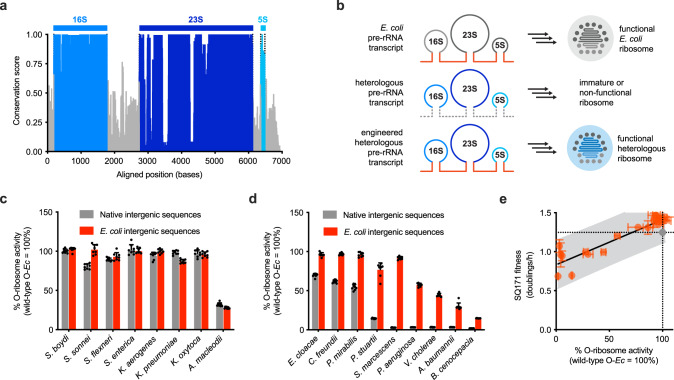
Fig. 3. Evaluating the effects of O-rRNA intergenic sequence replacement on heterologous translation.
a Per-base sequence conservation41 across 34 evaluated rRNA operons, showing limited conservation in intergenic regions as compared to structural rRNA genes. b Schematic representation of the intergenic sequence replacement strategy. c Effects of intergenic sequence replacement on O-rRNAs with high 16S rRNA sequence identity to E. coli (96.2–99.6%), as well as A. macleodii (85.9%), illustrating a minimal effect on orthogonal translation (n = 8). d Effects of intergenic sequence replacement on O-rRNAs with intermediate 16S rRNA sequence identity to E. coli (81.5–97.4%), illustrating a substantial effect on intergenic sequence replacement (n = 8). e Correlation between activity in the O-translation circuit and fitness in SQ171 strain complementation assays for 21 rRNAs evaluated after intergenic sequence replacement, showing a linear relationship. E. coli rRNA control plotted as a gray circle (99% CI, R2 = 0.84). Data reflect the mean and standard deviation of 3–8 biological replicates. Comprehensive SQ171 complementation and O-translation data (including precise number of replicates) reported in Supplementary Table 1. Wild-type O-Ec wild-type orthogonal E. coli rRNA, O-ribosome orthogonal ribosome, h hour. Source data are available in the Source Data File.