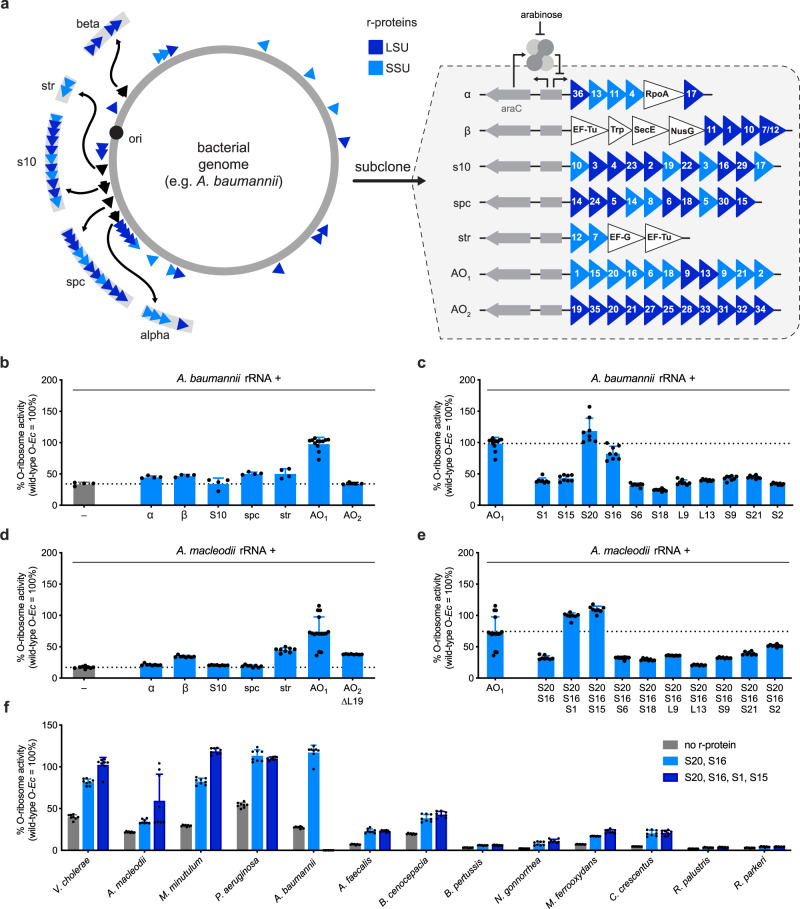
Fig. 4. Cognate r-proteins complementation improves heterologous O-rRNA activity.
a Schematic representation of natural ribosomal protein (r-protein) genomic organization for a given microbial genome and corresponding plasmid architecture for heterologous orthogonal ribosomal RNA (O-rRNA) complementation. b A. baumannii AO1 enhances cognate heterologous rRNA activity (n = 8 for AO1; otherwise n = 4). c A. baumannii S20 and S16 enhance A. baumannii O-ribosome activity to levels comparable to E. coli O-ribosomes (n = 8 for AO1; otherwise n = 4). d A. macleodii AO1 similarly improves cognate heterologous rRNA activity (n = 16 for AO1; otherwise n = 8). AO2 is expressed with L19 deleted due to observed toxicity (Supplementary Figs. 6a, b). e Cognate S1 or S15, alongside S20 and S16, maximize A. macleodii O-ribosome activity (n = 16 for AO1; otherwise n = 8). f Cognate S20, S16, S1, and S15 supplementation alongside cognate heterologous O-rRNAs. Toxicity is observed when expressing the four proteins together in A. macleodii and A. baumannii (Supplementary Fig. 7a; n = 8). Data reflect the mean and standard deviation of 4–16 biological replicates. Comprehensive O-translation data reported in Supplementary Table 1. Wild-type O-Ec wild-type orthogonal E. coli rRNA, SSU small subunit, LSU large subunit. Source data are available in the Source Data File.