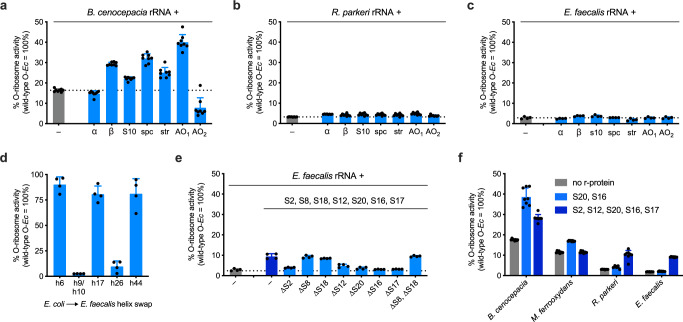
Fig. 5. Phylogenetically guided determination of cognate r-proteins that improve highly divergent heterologous O-ribosomes in E. coli.
a–c No single contiguous operon substantially improves translation activity of O-rRNAs derived from a B. cenocepacia (n = 8), b R. parkeri (n = 8), or c E. faecalis (n = 4). d Only 2 of the identified 5 regions with high sequence divergence between E. coli and E. faecalis 16S rRNAs abrogate E. coli O-translation when replaced with cognate E. faecalis sequences (n = 4). e Cognate S2, S8, S18, S12, S20, S16, and S17 (which directly contact h9, h10, and h2649) result in an increase in E. faecalis O-rRNA translation. Further analysis showed that S8 and S18 are not required for this increase (n = 4). f R-proteins S2, S12, S20, S16, and S17 collectively improve cognate O-rRNA translation for highly divergent species, but are not as effective as S20 and S16 alone for less divergent species. E. faecalis cognate ribosomal proteins (r-proteins) are expressed from a low copy number backbone (WT RepA SC101 origin) to limit toxicity (n = 8). Data reflect the mean and standard deviation of 4–8 biological replicates. Comprehensive O-translation data reported in Supplementary Table 1. Wild-type O-Ec wild-type orthogonal E. coli, O-ribosome orthogonal ribosome, h helix. Source data are available in the Source Data File.