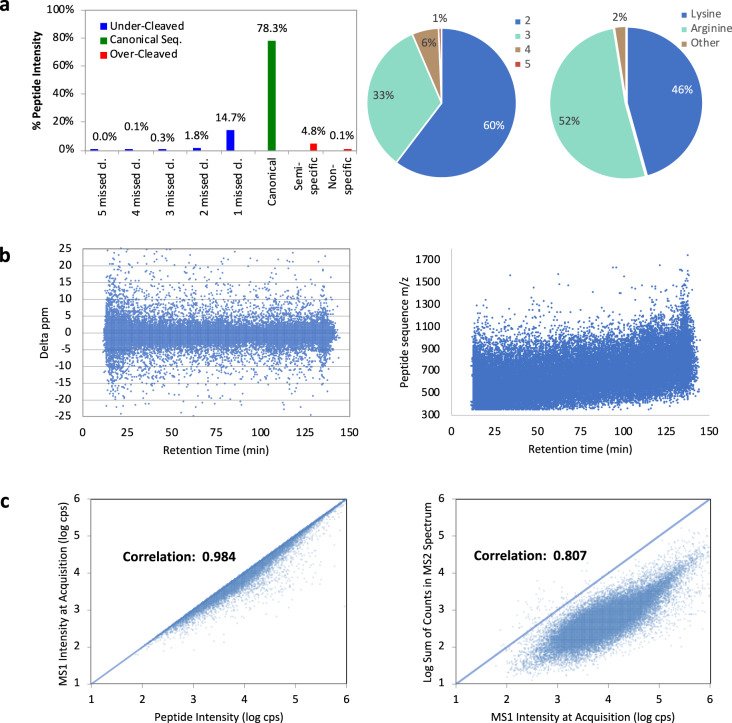
Fig. 3.
Statistical analysis of the proteomics data. (a) Enzymatic digestion efficiency with 78.2% of expected canonical sequence peptides, 5% over-cleaved sequence peptides, and 15.8% under-cleaved sequence peptides; the distribution of peptide precursor charge state with 60% doubly charged, decreasing to 5+ accordingly. The tryptic digestion resulted in uncontrolled C-terminal cleavage with 46% and 52% devoted to lysine and arginine respectively. (b) Precursor mass error in ppm during acquisition. The identified peptide sequence mass-to-charge value and its retention time in C18 chromatographic separation. (c) The correlation of MS1 intensity with good correlation to identified peptide intensity and good correlation between MS2 signals to MS1 intensity, showing the robustness of the mass spectrometry system in ion transmission and corresponding confidence on MS2 peptide mass fingerprint spectrum.