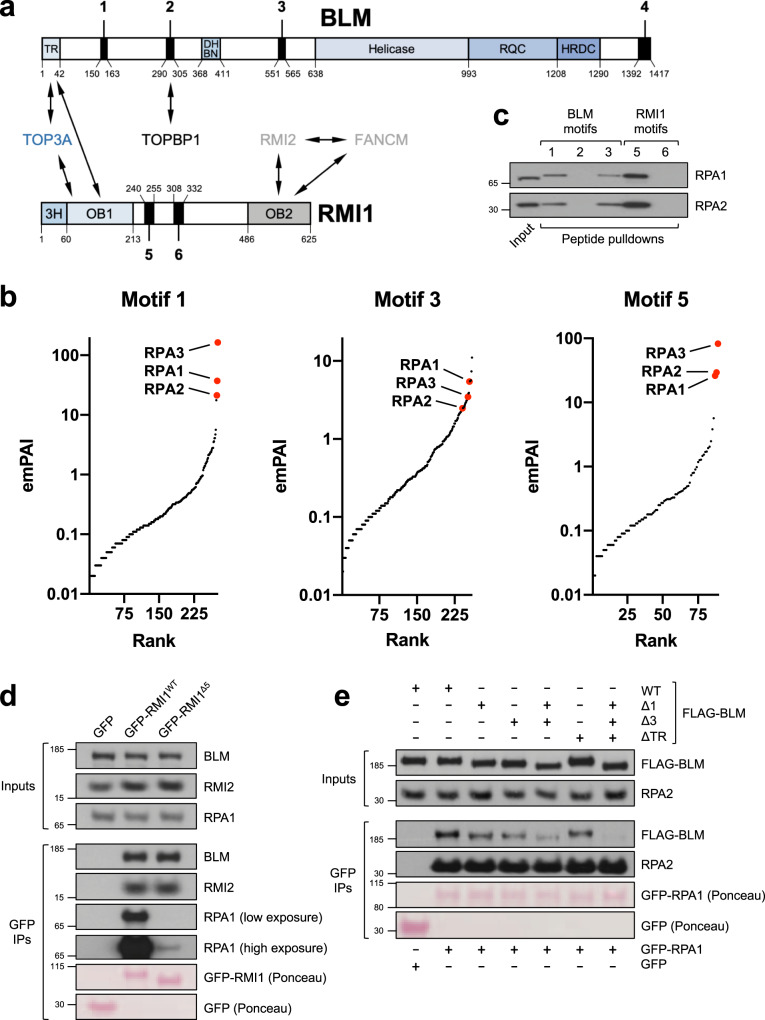
Fig. 1. Identification of conserved RPA-binding motifs in the BTR complex.
a Schematic of proteins, domains, and motifs in the BTR complex. TR TOP3A-RMI1-binding domain, DHBN dimerization helical bundle in N-terminal, RQC RecQ C-terminal, HRDC helicase and RNaseD C-terminal; 3H 3-helix bundle, OB oligonucleotide-binding. b Plots of MS hits from peptide pulldowns with motifs 1, 3, and 5, ranked by emPAI score116 (number of unique peptide sequences adjusted for protein size). RPA subunits are highlighted in red. c Validation of MS results by western blotting, showing that RPA from HeLa nuclear extracts is specifically pulled down by biotinylated peptides based on motifs 1, 3, and 5 but not motifs 2 and 6. d GFP-pulldowns from 293FT cells transfected with constructs expressing GFP or the indicated GFP-tagged RMI1 variants, showing that loss of motif 5 disrupts RPA-binding. e GFP-pulldowns from 293FT cells transfected with constructs expressing either GFP or GFP-RPA1, and the indicated FLAG-tagged BLM variants, showing how loss of motifs 1 and/or 3, and the N-terminal TOP3A-RMI1 (TR) binding domain of BLM impacts on RPA-binding.