Figure 4.
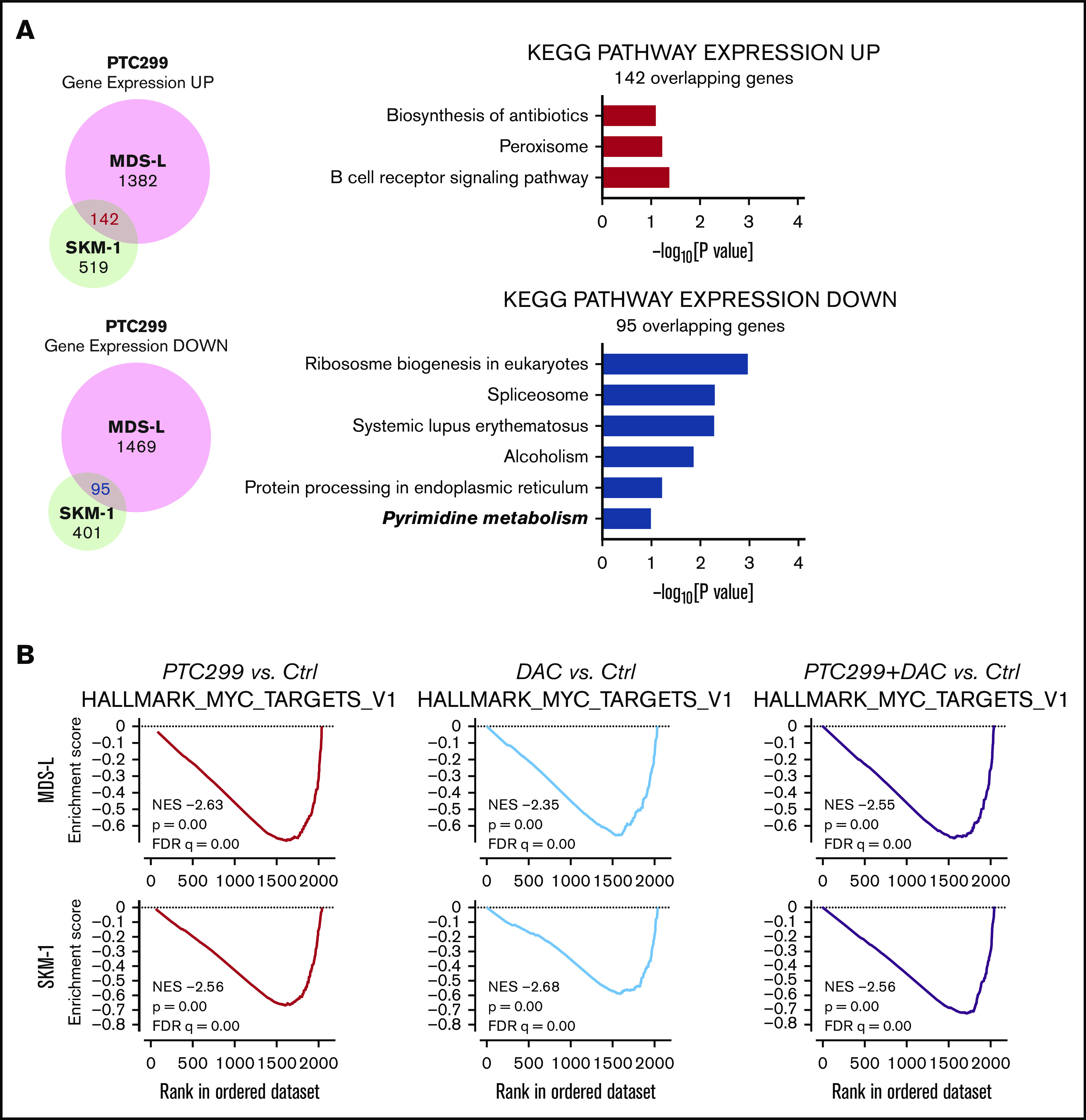
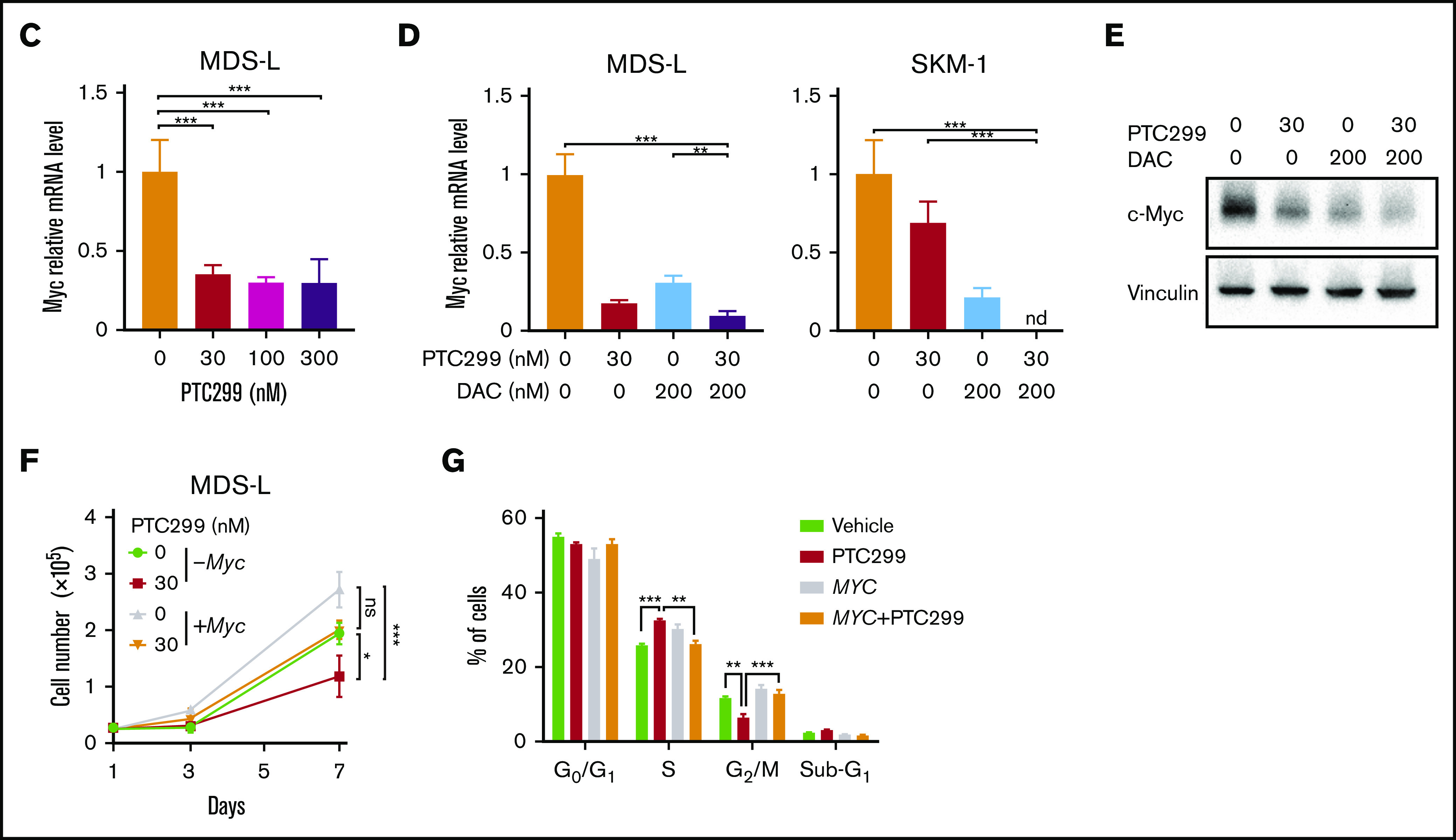
PTC299 downregulates c-MYC in MDS cells. (A) Venn diagram showing the overlap of down- (≤0.66-fold) or upregulated (≥1.5-fold) genes between MDS-L and SKM-1 cells treated with PTC299 relative to the DMSO control (left). Kyoto Encyclopedia of Genes and Genomes pathways enriched in overlapping genes (right) (B) GSEA plots showing the enrichment of MYC target genes in MDS-L (upper) and SKM-1 (lower) cells treated with PTC299 (left), DAC (center), and their combination (right) relative to control (Ctrl) cells. Normalized enrichment scores (NES), nominal P values (NOM), and false discovery rates (FDR) are indicated. (C-D) Quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) analysis of c-MYC in MDS cells treated with the indicated doses of (C) PTC299 and (D) 30 nM PTC299, 200 nM DAC as a single agent or in combination for 48 hours. GAPHD was used to normalize the amount of input RNA. nd, not detected. Data are shown as the mean ± SD (n = 3). (E) Immunoblotting of the extracts from MDS-L cells treated with PTC299 and/or DAC. (F) Growth of MDS-L cells with exogenous c-MYC in the presence of PTC299. MDS-L cells transduced with the Tet-on c-MYC plasmid were cultured in the presence and absence of 30 nM PTC299. c-MYC expression was induced by adding 1 μg/mL doxycycline. (G) The proportions of cells at the indicated phases of cell cycle are shown. MDS-L cells with or without exogenous c-MYC were treated with PTC299 for 72 hours. BrdU was added to the culture 2 hours before the analysis. Data are shown as the mean ± SD (n = 3). *P < .05, **P < .01, ***P < .001; ns, not significant by 1-way ANOVA.
