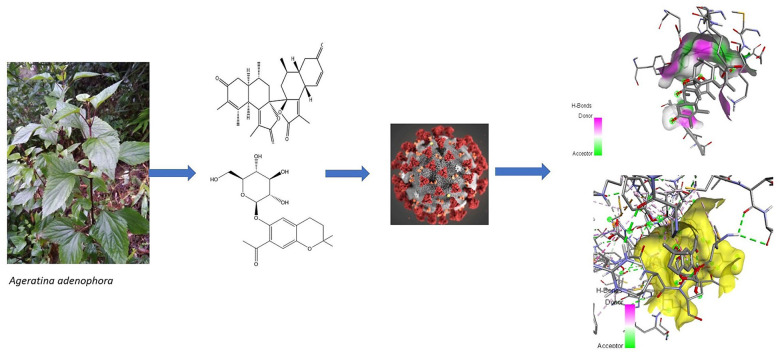
Abstract
The outbreak of COVID-19 created unprecedented strain in the healthcare system. Various research revealed that COVID-19 main protease (Mpro) and human angiotensin-converting enzyme 2 (ACE2) are responsible for viral replication and entry into the human body, respectively. Blocking the activity of these enzymes gives a potential therapeutic target for the COVID-19. The objective of the study was to explore phytochemicals from Ageratina adenophora against SARS-CoV-2 through in-silico studies. In this study, 34 phytochemicals of A. adenophora were docked with Mpro and ACE2 through AutoDock Tools-1.5.6 and their binding affinity was studied. Phytochemicals with higher affinity have been chosen for further molecular dynamics simulations to determine the stability with target protein. Molecular dynamics simulations were studied on GROMACS 5.1.4 version. Furthermore, 5-β-glucosyl-7-demethoxy-encecalin (5GDE) and 2-oxocadinan-3,6(11)-dien-12,7-olide (BODO) were found to be potential blockers with excellent binding affinity with Mpro and ACE2 than their native inhibitors remdesivir and hydroxychloroquine respectively. The drug likeness study and pharmacokinetics of the phytoconstituents present in A. adenophora provide an excellent support for the lead drug discovery against COVID-19.
Keywords: Ageratina adenophora, Angiotensin-converting enzyme, Main protease, Molecular docking, COVID-19
Graphical abstract
Abbreviations
- 5GDE
5-β-glucosyl-7-demethoxy-encecalin
- DAOA
2-deoxo-2-(acetyloxy)-9-oxoageraphorone
- OA
9-oxoagerophorone
- ODO
(+)-(5R,7S,9R,10S)-2-oxocandinan-3,6(11)-dien-12,7-olide
- BODO
(+)-7,7′-bis[(5R,7R,9R,10S)-2-oxocadinan-3,6(11)-dien-12,7-olide
- HEDO
(+)-(5R,7S,9R,10S)-7-hydroxy-7,12-epidioxycadinan-3,6(11)-dien-2-one
- CED
(−)-(5R,6R,7S,9R,10S)-cadinan-3-ene-6,7-diol
- 5-CQA
5-O-caffeoylquinic acid
- 3-CQA
3-O-caffeoylquinic acid
- 4-CQA
4-O-caffeoylquinic acid
- MPro
COVID-19 main protease
- ACE2
Human Angiotensin Converting Enzyme-2
- MDPC
methyl 3,4-bis[[(E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy]-1,5-dihydroxycyclohexane-1-carboxylate
- MDCQ
Methyl 3,5-di-O-caffeoyl quinate
1. Introduction
Coronavirus disease (COVID-19) caused by the novel coronavirus SARS-CoV-2 and declared as a global pandemic by WHO on March 11, 2020 (Ludwig and Zarbock, 2020; O’Horo, 2020). The first case of COVID-19 was reported in Wuhan, China in december (Tian et al., 2020). The virus is transmitted through direct or indirect contact of the respiratory droplet of an infected person and there is the possibility of air transmission (Brockmeier and Lager 2002; World Health Organization 2020). COVID-19 mainly affects the lungs and requires ventilator support in severe condition (NCT04382391, 2020). Signs and symptoms of infection include fever, dry cough, tiredness, ache and pain, sore throat, conjunctivitis, headache, loss of taste or smell, rash on the skin, or discoloration of fingers or toes (Sahai et al., 2020). SARS, MERS and SARS-CoV-2 belong to the same family, Coronaviridea (Chen et al., 2020). The outbreak SARS has infected 8096 and cause the death of 774 people (Bostanciklioglu 2020) and the MERS infect 2494 people and cause the death of 858 people. The recent outbreak of n-CoV-2 in November 2019, infect 62,686,326 and cause the death of 1,460,387 people globally till November 27, 2020. (Worldometers). The SARS-CoV-2 has a high mortality rate in the elder population, diabetes patients, people having cardiovascular diseases, and lungs diseases (Tskhay et al., 2020). Despite being so dangerous currently, there is no approved vaccine or drug for these viruses. Although hydroxychloroquine, lopinavir-ritonavir, favipiravir, remdesivir, tocilizumab are found effective in treating COVID-19 (Neupane et al., 2020)(Lu et al., 2020; Wu et al., 2020). Currently, several types of research and the clinical trial are going on worldwide to find the vaccine, but the success is still far away.
Structurally, SARS-CoV-2 consists of Spike glycoprotein (S), Envelope protein (E), Membrane protein (M), and Nucleocapsid protein (N). Coronaviruses bear positive-sense, single-stranded RNA (+ssRNA) genetic material with approximately 30,000 nucleotides, the longest identified genome in RNA viruses till date (Malik 2020). Viral genome of SARS-CoV-2 consists of 14 open reading frames (ORFs) (Bhosale et al., 2012). Among which the main reading frame ORF1ab encodes for two overlapping polyproteins 1a (pp1a) and replicase polyprotein 1 ab (pp1ab) (Cowley et al., 2000). These polyproteins play a major role in the replication and transcription of the viral genome (Robb et al., 2009). The virus has a protease class of hydrolytic enzyme known as Main protease (Mpro) also called as chymotrypsin-like protease (3CLpro). The entry of virus in the host has mediated binding Spike protein with human angiotensin-converting enzyme receptor-2 (ACE2) (Wan et al., 2020). Inside the host cell, proteolytic cleavage of pp1a and pp1ab by SARS-CoV-2 Mpro give rise to numerous non-structural proteins like RNA dependent RNA polymerase (RdRp), helicase and non-structural protein 3, 4, and 6 (nsp3, nsp4, and nsp6). These non-structural proteins are thought to be responsible for coronavirus replication. Thus, Mpro has emerged as important drug targets (Kirchdoerfer and Ward 2019) and as there is a very low similarity with human proteases and SARS-CoV-2 protease, inhibitors of Mpro will be very less toxic to humans. Human angiotensin-converting enzyme-2 (ACE2) is another potential target as it is directly involved in the entry of the virus into the host cell. ACE2 belongs to the angiotensin-converting enzyme family of dipeptidyl carboxydipeptidase. ACE2 protein catalyses the angiotensin I into angiotensin 1-9 and angiotensin II into the angiotensin 1-7. The ACE2 protein is a functional receptor for the spike glycoprotein of coronavirus diseases-2019 (COVID-19). (Magrone et al., 2020; Wan et al., 2020).
A. adenophora (commonly known as Crofton weed) is a perennial, semi-shrubby herbaceous plant which belongs to the Asteraceae family (Zhu et al., 2007). The plant is introduced as an ornamental plant, but leaf extract of the plant is found to have antimicrobial, antiseptic, analgesic, antipyretic, anti-inflammatory, larvicidal, insecticidal, wound healing, antitumor, and also antiviral activity (Poudel et al., 2020). Different research showed leaf extracts of A. adenophora have potential anti-viral activity against tobacco mosaic virus (TMV) and Human Immune virus (HIV). Jin et al. investigated antiviral activities of Eupatorium adenophorum leaf extract and it inactivated TMV particles and also effective in slight curative as well as preventive purpose (Jin et al., 2014). Ma et al. also suggested that A. adenophora has anti-HIV activity(MA et al., 2015). This excited us to do the in-silico screening of A. adenophora phytochemical in search of cheaper and less toxic phytochemicals against COVID-19 (André et al., 2019; Jin et al., 2014). Phytochemical screening reported that A. adenophora consist various class of secondary metabolites such as terpenoids, phenylpropanoids, flavonoids, coumarins, essential oils, sterols, phenolic acids, and alkaloids (Poudel et al., 2020; He et al., 2020). Among all the phytochemicals of A. adenophora, 34 phytochemicals were screened and docked with main protease (Mpro) of SARS-CoV-2 and angiotensin-converting enzyme receptor-2 (ACE2) of humans and results were compared with standard drugs which are found effective against SARS-CoV-2.
2. Methodology
2.1. Receptor preparation
The crystal structure of a COVID-19 main protease (Mpro) and human angiotensin-converting enzyme-2 (ACE2) related carboxypeptidase has been downloaded from protein data bank (PDB) website (http://www.rcsb.org/pdb)(Berman et al., 2000) (PDB ID: 6LU7 and 1R42) and were prepared by using Autodock tools-1.5.6. Kollman charges were added to each atom, non-polar hydrogens were merged, atom types were determined, and the structure of the prepared receptor were saved as pdbqt file.
2.2. Ligand preparation
The list of the major phytochemicals of A. adenophora were retrieved from the earlier reported literature (Poudel et al., 2020). The structures of known major chemical components of the plant were prepared using Chemdraw 3D pro12.0. software (Cousins 2005)and built - and - edit module in Open Babel 2.2.3 tool(O’Boyle et al., 2011). MMF F94s force field parameters were assigned to ligands atoms and then the energy has been minimized. Kollman charges were added to each ligand atoms. However, nonpolar hydrogen atoms were merged, and rotatable bonds defined. The structure of the ligands was saved in pdbqt format and used as in put ligand file for Autodock in docking.
2.3. Active site selection
The active site of the COVID-19 Mpro protein (PDB ID: 6LU7) and Human ACE2 protein (PDB ID: 1R42) has been predicted from different receptor cavities available. The different active site was identified from Discovery Studio 2020 (BIOVIA) client among these, site 1 of COVID-19 Mpro with coordinates (X = −10.711837, Y = 12.411388 and Z = 68.831286) and ACE2 receptor with coordinates (X = 44.98, Y = 20.98 and Z = 16.80) have been chosen as the active site.
2.4. Virtual molecular docking
The receptor and ligand interaction through virtual molecular docking have been performed through AutoDock Tools-1.5.6. All the molecules have been docked to active site 1 of COVID-19 Mpro and active site 1 of ACE2 receptor. Ligand-receptor interaction was viewed through Discovery Studio 2020 Client (BIOVIA) and re-docking of all the compounds was done to assure reproducibility (Pathak et al., 2020).
2.5. Molecular dynamic simulation
The effects of the ligands on the structural dynamics of proteins were studied on GROMACS 5.1.4 version (Abraham et al., 2015). Each complex and apo forms of the Mpro and ACE2 proteins were considered for the study. Additionally, the protein and water topology were defined by GROMOS43A2 force field (van Gunsteren et al., 1996) and SPC/E water model (Berendsen et al., 1987), respectively. Each ligand topology was generated through the same force field using the PRODRG server (http://prodrg2.dyndns.org/submit.html ). The charge of each simulating system was neutralized by an appropriate number of sodium ions (Na+). Subsequently, the energy of the simulating systems was minimized by the steepest descent algorithm in 50,000 steps. The two-phase equilibration of 100ps was carried out by NVT (at constant 300 K) and NPT (at constant 1 bar) ensemble equilibrations for each system. Consequently, MD simulation of 50ns was carried out for each system and the data was used for the enumeration of RMSD, RMSF and Rg. Furthermore, the binding energy was enumerated by the gMMPBSA package (Kumari et al., 2014) of GROMACS for every 0.1 ns frame of the last 20ns simulation of each protein-compound complex.
2.6. Drug likeness property
Physicochemical and pharmacokinetic properties to different phytochemical present in A. adenophora were studied using Swiss ADME a free online web tool (Zoete et al., 2016) (http://swissadme.ch/) by submitting their respective structures.
3. Result and discussion
3.1. Virtual screening through molecular docking
The SARS-CoV-2 Mpro with N3 inhibitor is divided into three domains. Domain I (from Phe 8 to Try 101), domain II (from Lys 102 to Pro 184) and domain III (from Thr 201 to Val 303). Domain I and II have an anti-parallel β-barrel structure. Domain III contains five α-helices arranged into the large anti-parallel globular cluster and is connected with domain II through loop region, which consist residue from Phe 185 to Ile 200. The substrate-binding site of Mpro of SARS-CoV-2 has Cys-His catalytic dyad, where N3 inhibitors bind. This is located in the cleft between domain I and domain II. Fig. 1 represents the crystal structure of COVID-19 main protease (Mpro) in complex with an inhibitor N3 and three domains of Mpro.
Fig. 1.
Crystal structure of COVID-19 main protease (Mpro) in complex with an inhibitor N3. Domain I shown in green, Domain II shown in cyan, Domain III is shown in red, and loop region shown in blue. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The validation of the docking calculation was evaluated by redocking Mpro inhibitor remdesivir on the main protease protein of SARS-CoV-2 (PDB ID: 6LU7). It displayed that remdesivir is able to bind in the almost same pose in the same binding pocket with a binding affinity of −7.8 kcal/mol. The study showed that remdesivir interacts with Mpro through various types of interaction with different amino acid residues by making hydrogen bond and Van der Waals bonds. However, it was noticed that the tested ligands interact with Glu A:166, Leu A:141, Gly A:143, Ser A:144, and Cys A:145 amino acids through hydrogen bonds.
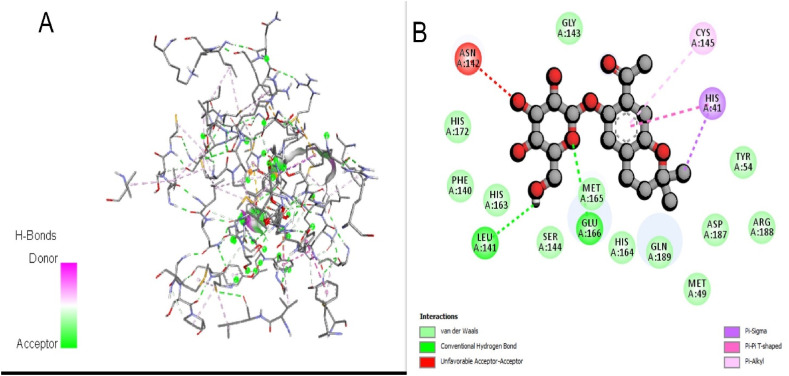
The interaction between phyto compound 5GDE and Mpro receptor is displayed in Fig. 2.
Fig. 2.
(A) The phytocompound 5GDE Mpro complex. (B) Detail amino acid interaction of 5GDE complex with Mpro in 2D.
These interactions are compared with native inhibitor N3 and it was found that 90.90% amino acid showed similarity with that of a co-crystal ligand, which was found acceptable. Thus, the protein was further utilized for docking 34 different compounds reported in A. adenophora and docking score is displayed in Table 1 (Poudel et al., 2020).
Table 1.
Docking score of 34 phytochemicals present in A. Adenophora with Main protease protein (Mpro) and human ACE2.
| S.N | Phytochemicals | Docking score of Mpro (affinity Kcal/mol) | Docking score of human ACE2 (affinity Kcal/mol) |
|---|---|---|---|
| 1 | 7-hydroxy-dehydrotremetone | -6.3 | -5.7 |
| 2 | 7,10,11-trihydroxy dehydrotremetone | -6.6 | -6.2 |
| 3 | 10-oxo-7-hydroxy-nordehydrotremetone | -6.2 | -5.7 |
| 4 | 2αmethoxyl-3β-methyl-6-(acetyl-O-methyl)-2,3-dihydrobenzofuran | -5.7 | -5.3 |
| 5 | 5-b-glucosyl-7-demethoxy-encecalin (5GDE) | -8.1 | -6.5 |
| 6 | 8-hydroxy-8-b-glucosyl-2-carene | -6.6 | -5.7 |
| 7 | (4S,4aR,6R)-1-acetyl-6-(acetyloxy)-4,4a,5,6-tetrahydro-4,7-dimethylnaphthalen-2(3H)-one | -6.6 | -5.7 |
| 8 | 2-deoxo-2-(acetyloxy)-9-oxoageraphorone(DAOA) | -6.2 | -6.0 |
| 9 | 9-oxoagerophorone(OA) | -5.9 | -5.3 |
| 10 | 9-oxo-10,11-dehydro-agerophorone(ODA) | -6.0 | -5.9 |
| 11 | 9β-hydroxy-ageraphorone | -5.8 | -5.4 |
| 12 | Muurol-4-en-7-ol | -5.7 | -5.5 |
| 13 | 8-beta-hydroxy-9,12-dehydroverbocciolenten | -5.5 | -5.2 |
| 14 | 2 β-acetoxy-(7α, 9β H)-3.6(11)-cadinadien-12(7)-olide | -6.9 | -6.4 |
| 15 | 3-hydroxymuurola-4,7 (11)- dien-8-one | -6.1 | -5.5 |
| 16 | (+)-(5R,7S,9R,10S)-2-oxocandinan-3,6(11)-dien-12,7-olide (ODO) | -6.8 | -6.5 |
| 17 | (+)-7,7′-bis[(5R,7R,9R,10S)-2-oxocadinan-3,6(11)-dien-12,7-olide (BODO) (He et al., 2008) | -6.3 | -8.8 |
| 18 | (+)-(5R,7S,9R,10S)-7-hydroxy-7,12-epidioxycadinan-3,6(11)-dien-2-one (HEDO) | -6.7 | -6.3 |
| 19 | (−)-(5R,6R,7S,9R,10S)-cadinan-3-ene-6,7-diol (CED) | -5.8 | -5.5 |
| 20 | (+)-(5S*,6R*,9R*,10S*)-5,6-dihydroxycandinan-3-ene-2,7-dionel | -6.0 | -5.7 |
| 21 | 7-hydroxycandinan-3-ene-2-one | -5.6 | -5.4 |
| 22 | 5,6-dihydroxy candinan-3-ene-2,7-dione | -6.5 | -5.9 |
| 23 | 2-acetyl-candinan-3,6-diene-7-one | -6.1 | -6.1 |
| 24 | Candinan-3-ene-2,7-dione | -5.8 | -5.3 |
| 25 | Candinan-3,6-diene-2,7-dione | -6.2 | -5.6 |
| 26 | 1,6-dihydroxy-1-isopropyl-4,7-dimethyl-3,4dihydronaphthalen-2(1H)-one | -6.4 | -5.8 |
| 27 | (4R,5S)-4-Hydroxy-5-isopropyl-2-methyl-2-cyclohexehone | -4.9 | -4.6 |
| 28 | 5-O-caffeoylquinic acid (5-CQA) | -7.2 | -6.3 |
| 29 | 3-O-caffeoylquinic acid (3-CQA) | -8.0 | -6.4 |
| 30 | 4-O-caffeoylquinic acid (4-CQA) | -7.6 | -6.4 |
| 31 | 5-O-trans-o-coumaroylquinic acid methyl ester | -7.0 | -6.3 |
| 32 | methyl (1R,3S,4S,5S)-3-[(E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy-1,4,5-trihydroxycyclohexane-1-carboxylate | -7.3 | -6.2 |
| 33 | methyl 3,4-bis[[(E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy]-1,5-dihydroxycyclohexane-1-carboxylate (MDPC) | -7.4 | -7.3 |
| 34 | Methyl 3,5-di-O-caffeoyl quinate (MDCQ) | -8.0 | -6.8 |
It was found that among these only three compounds (5GDE, 3-CQA and MDCQ) have better binding affinities than remdesivir. Moreover, 5GDE showed the highest docking affinity (−8.1 kcal/mol) in the entire set of molecules. The comparison between remdesivir and phytocompound 5GDE complex displayed 100% similarity in interaction pattern, illustrated in Table 2 . Among all the interacting amino acids of 3-CQA and MDCQ it was found that 94.11% and 94.73% amino acid interactions are similar with remdesivir, respectively. The docking score along with hydrogen bond and Van der Waals interactions of these three compounds with MPro is shown in Table 2.
Table 2.
Molecular interaction of top 3 high scoring compounds with Main protease protein (Mpro) of SARS-CoV-2.
| S.N. | Compounds | H-bond interactions | Van der waals interactions |
|---|---|---|---|
| 1. | 5GDE | LEU A:141, GLU A: 166 | MET A:49, TRY A:54, ARG A:188, ASP A:187, GLN A:189, SER A:144, MET A:165, SER A:144, HIS A:163, PHE A:140, HIS A:172, GLY A:143 |
| 2. | 3-CQA | GLU A: 166, CYS A: 145, THR A: 190 | HIS A:172, PHE A:140, LEU A:141, ASN A:142, SER A:144, GLN A:189, ARG A:188, GLN A:192, ALA A:191, PRO A:168, HIS A:164 |
| 3 | MDCQ | LEU A:141, HIS A:163, SER A:144, SER A:144, GLN A:192 | PHE A:140, ASN A:142, GLY A:143 TRY A:54, ASP A:187, ARG A:188, GLN A:189, THR A: 190, LEU A:167, GLN A: 166 |
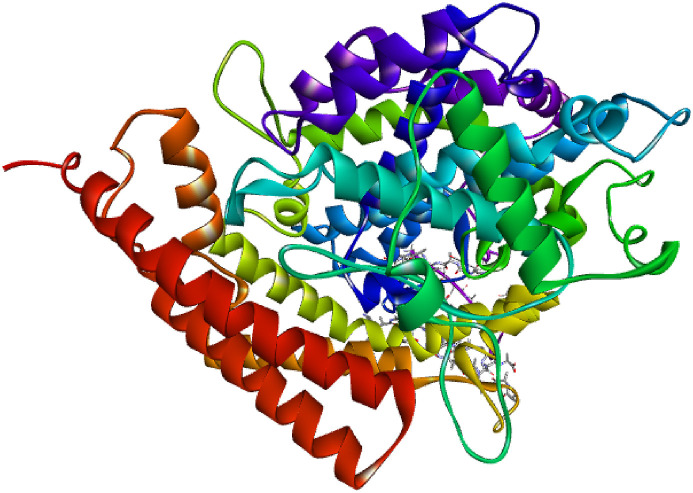
The human ACE2 enzyme is composed of two domains; the first is zinc metallopeptidase domain depicted in Fig. 3 (residues 19–611) and the second C terminus domain (residues 612–740). The first domain is 42% identical to the corresponding domains of humans ACE. The first zinc metallopeptidase domain is located near the bottom and on one side of the large active site cleft. The zinc is coordinated by His374, His 378, Glu402 and single water molecules (in the native structure). The second domain is identical to human collectrin by 48%. The ACE2 metallopeptidase domain was divided into two subdomains (I and II), that formed the two sides of a long deep cleft with dimensions of 40 Ᾰ long by 15Ᾰ wide by ~25Ᾰ deep. The catalytic subdomains are only connected at the floor of the active site cleft.
Fig. 3.
The crystal structure of COVID-19 zinc metallopeptidase domain of ACE2 (PDB ID: 1R42). Surface is created around the co-crystal ligand present in the binding pocket in which docking is performed for the study.
Initially, re-docking of the endogenous or native co-crystal ligand has been performed for the validation of the whole docking procedure ensuring its reproducibility. A total of 34 phytochemicals of A. adenophora have been docked with human ACE2. The receptor-ligand interactions have indicated that all the compounds used for docking procedure have a substantial binding affinity among them top 5 high scoring compounds are listed in Table 3 .
Table 3.
Enlisting of molecular interactions of top 5 high scoring A. adenophora phytochemical with human ACE2 protein.
| S.N. | Compound Name | Hydrogen bond | Van der Waals interaction |
|---|---|---|---|
| 1 | BODO | - | UNK C:909, PRO A: 492, TYR A:613, GLU A:495, GLU A:489, TRP A: 478, HIS A: 493, ASP A:494, LYS A:475, UNK C:907, MET A:474 |
| 2 | MDPC | TYR A:613, ARG A:482, ASP A:471, GLU A:489 | UNK C:910, UNK C:909, ASP A:615, SER A:611, ASP A:609, ASP A:494, HIS A:493, GLN A:472, TRP A:478 |
| 3 | 5GDE | HIS A:493 | - |
| 4 | ODO | ARG A:482 | TRP A:478, PRO A:492, GLU A:489, THR A:608 |
| 5 | MDCQ | HIS A:493, GLU A:479 | SER A:611, ALA A:614, TYR A:613, UNK C:909, ARG A:482, TRP A:478, UNK C:908, UNK C:907, MET A:474, ASP A:494 |
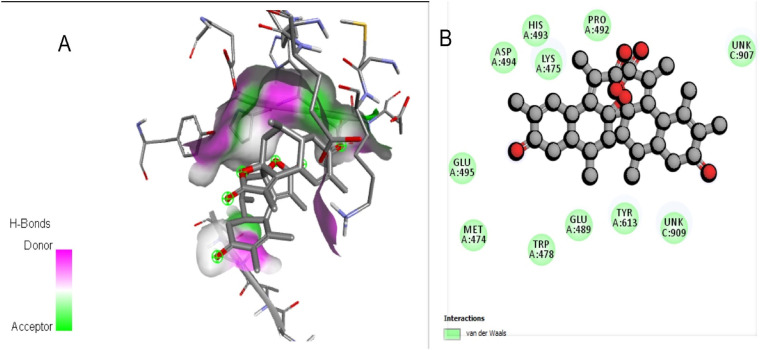
Top 5 high scoring compounds has been taken into consideration for further detailed analysis as reflected in Table 3. BODO has shown the best binding affinity towards the human ACE2 receptor with a binding energy of −8.7 kcal/mol. This binding energy is higher than best fit inhibitor (Hydroxychloroquine) binding affinity, which is −6.3 kcal/mol. The best fit inhibitor was selected by comparing binding affinity of several drugs that have shown activity against COVID-19. The interaction of the compound with the human ACE2 receptor is depicted in Fig. 4 . Docking study has exhibited that BODO interacts with 11 amino acid residue among which 7 residues (GLU A:495, ASP A:494, PRO A:492, HIS A:493, TRP A:478, TYR A:613, GLUA:489) matches with that to the inhibitor hydroxychloroquine. Thus, can be predicted compound binds to the receptor properly to inhibit human ACE2 protein.
Fig. 4.
(A) The phytocompound BODO ACE2 complex. (B) Detail amino acid interaction of BODO with ACE2 receptor in 2D.
MDPC, a phytochemical of A. adenophora, has shown a binding energy score of −7.5 kcal/mol, which is higher than the native inhibitor. The study stated that MDPC interacted with 10 amino acid residues (TYR A:613, ARG A:482, GLU A:489, ASP A:494, ASP A:471, HIS A:493, TRP A:478, GLN A:472, GLU A:495 and GLU A:479), which matches with the hydroxychloroquine. Further, it was also noticed that 5GDE have higher binding affinity (−6.7 kcal/mol) with human ACE2 than native inhibitor hydroxychloroquine. Additionally, the careful study of interaction emphasized that the compound 5GDE interacts with two amino acids through hydrogen and pi-alkyl bonds, where one amino acid residue HIS A: 493 found common to the native hydroxychloroquine.
A. adenophora phytochemical have already proven anti-TMV and anti-HIV activity. In our in-silico study molecular dynamics of phytochemicals of A. adenophora done with Mpro and ACE2 virtually and found that phytochemicals BODO and 5GDE, were potential blockers of Mpro and ACE2. These plant materials are extremely abundant, convenient to harvest and easy to collect. This will lead to cost effective drug can be develop.
3.2. Molecular dynamic simulation
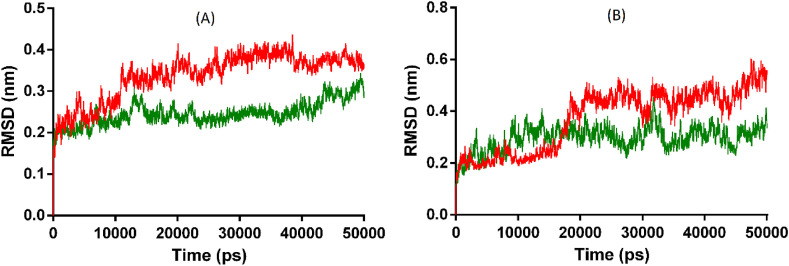
To elucidate the effects of the selected compounds (BODO and 5GDE) on the structural attributes of the respective protein (ACE2 and Mpro), the comparative MD analysis was carried out between each protein complex and its apo form. The average RMSD of Mpro complex and apo form was enumerated as 0.37, and 0.29 nm, wherein Mpro exhibited low deviations in backbone atoms with 5GDE in comparison to the apo structure. The RMSD graph is displayed in Fig. 5 .
Fig. 5.
A and B displayed RMSD of ACE2 and Mpro.
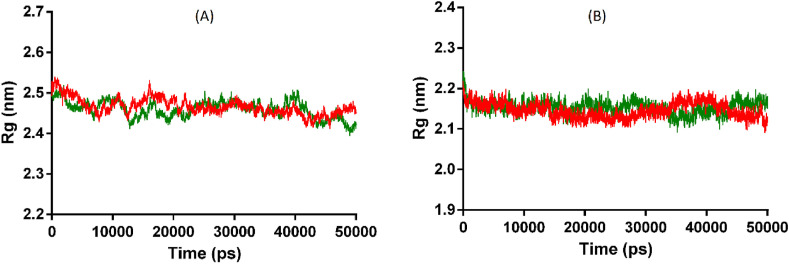
Further, the effect of interaction between compound and Mpro protein on its compactness was analyzed by Rg analysis. The respective average Rg values were computed as 2.14 and 2.15 nm for apo and complex form, suggesting that both forms exhibited a similar level of compactness during simulation. The backbone atom fluctuation of the Mpro residues in both forms was analyzed by the RMSF data, which showed that apo and complex form have an average RMSF of 0.21and 0.15 nm, respectively. It indicates that backbone atoms of Mpro residues showed low fluctuation in complex form in comparison to the apo form. The MD analysis showed that the Mpro complex structure retains similar compactness like its apo structure, but with increased structural stability and low residue fluctuation in comparison to the apo form during the simulation. Analysis also depicts the stability of interactions between 5-β-glucosyl-7-demethoxy-encecalinand protein during the dynamics. Simultaneously, the average RMSD of the respectiveACE2 complex and apo form was enumerated as 0.34, and 0.25 nm that delineate lesser deviations in the backbone atoms of the ACE2-compound complex in comparison to its apo structure. The effect on compactness of the ACE2 after accommodating the BODO was analyzed through Rg analysis. The average Rg value for the ACE2 apo and complex form was found to be 2.47 and 2.46 nm, respectively, indicating that both apo and complex forms of ACE2 protein attained a similar level of compactness. The Rg graph is displayed in Fig. 6 .
Fig. 6.
(A) and (B) displayed Rg of ACE2 and Mpro.
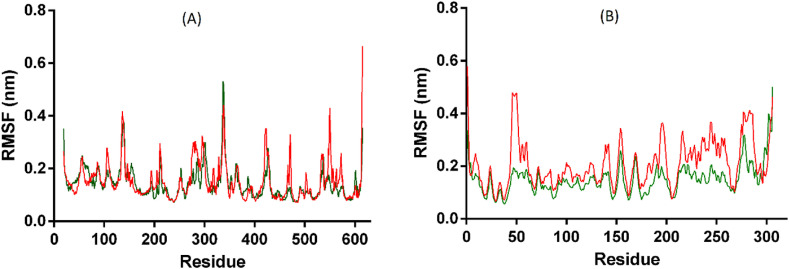
The backbone atom fluctuation of the ACE2 residues was analyzed by the RMSF data, which showed similar levels of fluctuation in apo and complex forms with an average RMSF of 0.15 and 0.14 nm, respectively. The MD analysis showed that the ACE2 complex structure retains similar compactness and residues motion like its apo structure, but with increased structural stability during the simulation. The RMSF graph is displayed in Fig. 7 .
Fig. 7.
(A) and (B) displayed RMSF of ACE2 and Mpro.
In addition, the interface affinity of the protein and compound was analyzed by calculating the binding energy of the protein complex. The average binding energy calculated for ACE2 (BODO) and Mpro-(5GDE) complex is presented in Table 4 which represents that Van der Waals energy played a major role in maintaining the binding of compounds with protein in both of the complexes during the dynamics.
Table 4.
The Binding energy of the protein-compound complex. Drug likeness property.
| Complex | Van der Waal energy (Kj/mol) | Electrostatic energy (Kj/mol) | Polar salvation energy(Kj/mol) | SASA energy(Kj/mol) | Binding energy (Kj/mol) |
|---|---|---|---|---|---|
| ACE2-Comp | -181.67 ± 0.97 | -7.39 ± 0.62 | 98.33 ± 1.51 | -15.75 ± 0.08 | -106.52 ± 1.33 |
| Mpro-Comp | -166.48 ± 0.82 | -40.96 ± 0.65 | 108.23 ± 0.89 | -17.03 ± 0.074 | -116.31 ± 0.89 |
Drug likeness is a crucial step in the initial step of drug discovery. The pharmacological properties of 4 high scoring phytochemicals of A. adenophora were studied. The pharmacokinetics profiling and toxicity predictions need to assess its drug-likeness property using Swiss ADME, which works on interpreting the molecular fingerprint of the submitted query structure, and analyze the presence or absence of chemical features in a molecule to publish pharmacokinetic data. The in silico pharmacokinetic predictions of the top 10 compounds of A. adenophora are depicted in Table 5 .
Table 5.
In silico pharmacokinetics prediction (Lipinski parameters) for the top 4 phytochemicals of A. adenophora.
| Pharmacological properties | BODO | 5GDE | MDCQ | MDPC |
|---|---|---|---|---|
| MW | 504.61 | 382.40 | 532.49 | 516.49 |
| n-rotb | 1 | 4 | 11 | 10 |
| n-HbA | 6 | 8 | 12 | 11 |
| n-HbD | 0 | 4 | 6 | 6 |
| ILog P | 3.12 | 2.45 | 2.92 | 2.39 |
| Lipinski violations | 1 | 0 | 3 | 3 |
| Lead likeness violations | 1 | 1 | 2 | 2 |
MW: Molecular Weight, n-rotb: Number of rotable bonds, n-HbA: Number of hydrogen bond acceptor, n-HbD: Number of hydrogen bond donors.
Four compounds depicted in the table are high scoring compounds in 2 different receptors ACE2 and Mpro but BODO and 5GDE are the top-scoring compounds, respectively. These compounds also have single lead likeness violations. Therefore, these molecules can be considered for the further drug development steps. Among these two 5GDE has a molecular weight less than 500, which follow Lipinski's rule of five. This can propound that the compounds are safer and permissible for further study. The computational activity predication for expressed that selected phyto-compounds have mild to moderate potency against COVID-19 proteins.
3.3. Probable mechanism of action
The coronavirus is positive-strand RNA viruses (Weiss and Navas-Martin 2005). The process of viral replication and transcription is controlled by two overlapping polyproteins, the pp1a (replicase 1a, 450KD) and pp1ab (replicase 1 ab, 750KD) (Yang et al., 2003). Frame shifting of the ribosome is essential for the expression of the C-proximal of pp1ab polyprotein (Kjær and Belsham 2018). The release of functional polypeptides from both polyproteins is controlled by Mpro, and necessary for proteolytic processing of polyproteins translated from viral RNA. The main protease slices the polyprotein by autolytic cleavage of the enzymes. This indicates blocking the activity of the main proteases could lead to inhibition of viral replication 5GDE, inhibits the Mpro with more binding affinity than native inhibitor through the multiple types of interactions.
Coronavirus also presents on protein S a specific binding site for ACE2 which serves as an entry point into the host cell. Various researches on ACE2 suggest, blocking the ACE2 receptor could make it more difficult for coronavirus to enter cells. BODO binds to the ACE2 receptor more effectively with higher binding affinity and most of the amino acid residue (GLU A:495, ASP A:494, PRO A:492, HIS A:493, TRP A:478, TYR A:613, GLUA:489) matches with native inhibitor hydroxychloroquine. 5GDE and BODO are phytochemicals of A. adenophora inhibit the viral replication and block the entry of SARS-CoV-2 through Mpro and ACE2 receptors. Phytochemicals of these two A. adenophora phytochemicals have shown more effective binding energy than its native inhibitors remdesivir and hydroxychloroquine.
4. Conclusion
Our aim of the study was to investigate efficiency of the phytocompounds against coronavirus. Therefore, the phytochemicals of A. adenophora are in-silico tested against Mpro and ACE2 receptor. Furthermore, the docking calculation confirmed that the phytocompound 5GDE consist significant identical interaction with selected protein as referenced compounds. However, the binding energy was measure slightly above than standard drug remdisivir and hydroxychloroquine. The MD analysis of Mpro complex structure found to retain similar compactness like its apo structure, but with increased structural stability and low residues fluctuation in comparison to the apo form during the simulation. Whereas, phytochemical BODO has shown high binding affinity (score), almost stable interaction with ACE2 receptor, retain similar compactness, and residues motion like its apo structure, but with increased structural stability during the simulation.
Our study showed natural compounds from A. adenophora have either similar or greater binding affinity than that of natural inhibitor remdesivir and hydroxychloroquine. Hence, 5GDE and BODO can be further developed as drug candidates against Mpro and ACE2 receptor of coronavirus, responsible for ongoing pandemic. Finally, we suggest from this in-silico study that A. adenophora phytochemicals block both Mpro and ACE2 receptors.
Declaration of competing interest
Authors have no conflict of interest regarding this article.
Acknowledgements
The authors are very grateful to the Department of Biotechnology and Bioinformatics, School of Life Sciences, University of Hyderabad, Hyderabad, Telangana, India. Prateek Pathak acknowledged that the work was supported by Government of the Russian Federation, contract 02. A03.21.0011.
References
- André R., Catarro J., Freitas D., Pacheco R., Oliveira M.C., Serralheiro M.L., Falé P.L. Action of euptox A from Ageratina adenophora juice on human cell lines: a top- down study using FTIR spectroscopy and protein profiling. Toxicol. Vitro. 2019;57:217–225. doi: 10.1016/j.tiv.2019.03.012. [DOI] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhosale U.A., Yegnanarayan R., Pophale P., Somani R. Effect of aqueous extracts of Achyranthes aspera Linn. on experimental animal model for inflammation. Ancient Sci. Life. 2012;31:202–206. doi: 10.4103/0257-7941.107362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostanciklioglu S.M., Temiz E. Severe acute respiratory syndrome coronavirus 2 is penetrating to dementia research. Curr. Neurovascular Res. 2020:1–5. doi: 10.2174/1567202617666200522220509. [DOI] [PubMed] [Google Scholar]
- Brockmeier S.L., Lager K.M. Experimental airborne transmission of porcine reproductive and respiratory syndrome virus and Bordetella bronchiseptica. Vet. Microbiol. 2002;89(4):267–275. doi: 10.1016/s0378-1135(02)00204-3. [DOI] [PubMed] [Google Scholar]
- Chen Z.L., Zhang W.J., Lu Y., Guo C., Guo Z.M., Liao C.H., Zhang X., Zhang Y., Han X.H., Li Q.L., Lu J.H. From SARS-CoV to 2019-nCoV outbreak: similarities in the early epidemics and prediction of future trends. Chinese Med J. 2020;133(9):1112–1114. doi: 10.1097/CM9.0000000000000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins R.K. ChemDraw ultra 9.0. CambridgeSoft, 100 CambridgePark drive. J. Am. Chem. Soc. 2005;127(11):4115–4116. doi: 10.1021/ja0410237. www.cambridgesoft.com Cambridge, MA 02140. [DOI] [Google Scholar]
- Cowley J.A., Dimmock C.M., Spann K.M., Walker P.J. Gill-associated virus of Penaeus monodon prawns: an invertebrate virus with ORF1a and ORF1b genes related to arteri-and coronaviruses. J. Gen. Virol. 2000;81(6):1473–1484. doi: 10.1099/0022-1317-81-6-1473. [DOI] [PubMed] [Google Scholar]
- He L., Hou J., Gan M., et al. Cadinane sesquiterpenes from the leaves of Eupatorium adenophorum. J. Nat. Prod. 2008;71(8):1485–1488. doi: 10.1021/np800242w. [DOI] [PubMed] [Google Scholar]
- Jin Y., Hou L., Zhang M., Tian Z., Cao A., Xie X. Antiviral activity of Eupatorium adenophorum leaf extract against tobacco mosaic virus. Crop Protect. 2014;60:28–33. [Google Scholar]
- Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10(1):1–9. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjær J., Belsham G.J. Modifications to the foot-and-mouth disease virus 2A peptide: influence on polyprotein processing and virus replication. J. Virol. 2018;92(8) doi: 10.1128/JVI.02218-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C.C., Chen M.Y., Lee W.S., Chang Y.L. Potential therapeutic agents against COVID-19: what we know so far. J. Chin. Med. Assoc. 2020;1–3 doi: 10.1097/JCMA.0000000000000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q.P., Cheng C.R., Li X.F., Liang X.Y., Ding J. Chemistry, pharmacological activities and analysis of Ageratina adenophora. Asian J. Chem. 2015;27:4311–4316. [Google Scholar]
- Magrone T., Magrone M., Jirillo E. Focus on receptors for coronaviruses with special reference to angiotensin-converting enzyme 2 as a potential drug target-a perspective. Endocrine, Metabolic & Immune Disorders-Drug Targets (Formerly Current Drug Targets-Immune. Endocrine & Metabolic Disorders) 2020;20(6):807–811. doi: 10.2174/1871530320666200427112902. [DOI] [PubMed] [Google Scholar]
- Malik Y.A. Properties of coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020;42(1):3–11. [PubMed] [Google Scholar]
- NCT04382391 Study assessing vagus nerve stimulation in covid-19 respiratory symptoms. 2020. https://clinicaltrials.gov/show/NCT04382391
- Neupane N.P., Das A.K., Singh A.K., Verma A. Off label medication to combat COVID-19: review Results to Date. Bentham Sci Coronaviruses. 2020;1:1–2. [Google Scholar]
- O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: an open chemical toolbox. J. Cheminf. 2011;3(1):33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak P., Novak J., Naumovich V., Grishina M., Balkrishna A., Sharma N., Sharma V., Potemkin V., Verma A. Polyphenolic rich extract of Oroxylum indicum alleviate β-glucuronidase activity via down-regulate oxidative stress: experimental and computational studies. Biocatalysis and Agricultural Biotechnology. 2020;29:101804. [Google Scholar]
- Poudel R., Neupane N.P., Mukeri I.H., Alok S., Verma A. Vol. 11. 2020. An updated review on invasive nature, phytochemical evaluation, & pharmacological activity of Ageratina Adenophora; pp. 2510–2520. 6. [DOI] [Google Scholar]
- Robb N.C., Smith M., Vreede F.T., Fodor E. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J. Gen. Virol. 2009;90(6):1398–1407. doi: 10.1099/vir.0.009639-0. [DOI] [PubMed] [Google Scholar]
- Sahai R., Dutta S., Kumar T. Anticoagulants in COVID-19 therapy: an evidence-based review. Int. J. Pharmaceut. Sci. Rev. Res. 2020;63(1):191–195. [Google Scholar]
- Tian H., Liu Y., Li Y., Wu C.H., Chen B., Kraemer M.U., Li B., Cai J., Xu B., Yang Q., Wang B. An investigation of transmission control measures during the first 50 days of the COVID-19 epidemic in China. Science. 2020;368(6491):638–642. doi: 10.1126/science.abb6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tskhay A., Yezhova A., Alibek K. COVID-19 pandemic: is chronic inflammation a major cause of death? medRxiv. 2020:1–16. doi: 10.1101/2020.05.12.20099572. [DOI] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94(7):1–9. doi: 10.1128/jvi.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss Susan R. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus/susan R. Weiss, sonia navas-martin. Microbiol. Mol. Biol. Rev. 2005;69(4):635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; 2020. Modes of Transmission of Virus Causing COVID-19: Implications for IPC Precaution Recommendations: Scientific Brief; p. 27. March 2020 (No. WHO/2019-nCoV/Sci_Brief/Transmission_modes/2020.1. [Google Scholar]
- Worldometers Coronavirus cases. 2020. https://www.worldometers.info/coronavirus/ Retrieved November 27.
- Wu R., Wang L., Kuo H.C.D., Shannar A., Peter R., Chou P.J., Li S., Hudlikar R., Liu X., Liu Z., Poiani G.J. An update on current therapeutic drugs treating COVID-19. Current Pharmacology Reports. 2020:1. doi: 10.1007/s40495-020-00216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. Unit. States Am. 2003;100(23):13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Sun O.J., Sang W., Li Z., Ma K. Predicting the spatial distribution of an invasive plant species (Eupatorium adenophorum) in China. Landsc. Ecol. 2007;22(8):1143–1154. [Google Scholar]
- Zoete V., Daina A., Bovigny C., Michielin O. Swiss similarity: a web tool for low to ultra high throughput ligand-based virtual screening. J. Chem. Inf. Model. 2016;56(8):1399–1404. doi: 10.1021/acs.jcim.6b00174. [DOI] [PubMed] [Google Scholar]