Figure 3.
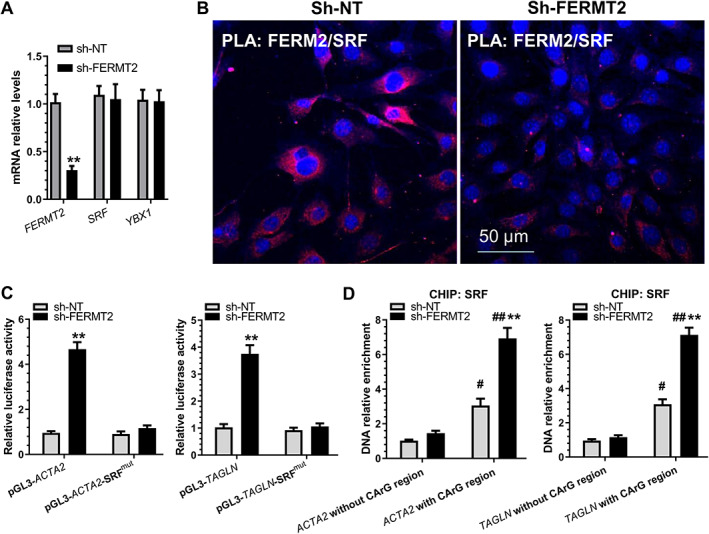
FERM2 suppresses SMC gene expression by interacting with and retaining SRF within cytoplasm. HUVECs infected with a non‐target (sh‐NT) or FERMT2 gene‐specific shRNA (sh‐FERMT2) lentivirus were incubated with 5 ng/ml TGFβ1/TNFα for 4 days to induce EndoMT. (A) RT‐qPCR analysis of gene expression. (B) Proximity ligation assays (PLAs) using a pair of primary antibodies (or respective IgG controls) as indicated to detect the in situ protein interactions of FERM2 with SRF in HUVECs undergoing EndoMT. (C) SMC gene promoter activity assays showed that SRF binding sites(s) are required for FERM2‐mediated SMC gene suppression. HUVECs infected with sh‐NT or sh‐FERM2 lentivirus were transfected with wild‐type SMC gene promoters (pGL3‐ACTA2/TAGLN) or their SRF binding site mutants (pGL3‐ACTA2/TAGLN‐SRFmut), respectively. Luciferase activity assay was conducted at 48 h post‐transfection. (D) FERMT2 knockdown increased the binding and enrichment of SRF to SMC gene promoters. ChIP assays were performed using antibody against SRF or normal IgG, respectively, as described in the Materials and methods section. Quantitative PCR amplifications of the adjacent regions without SRF binding sites (CArG element) were included as an additional control for specific promoter DNA enrichment. The data presented here are representative images (B) or mean ± SEM (A, C, D) of five (n = 5) independent experiments. **p < 0.05 (versus sh‐NT); # p < 0.05, ## p < 0.01 (versus promoter DNA without CArG region). Student's t‐test (A) or two‐way ANOVA with a Tukey's post hoc test (C, D) was used for statistical analysis, respectively.
