Figure 3.
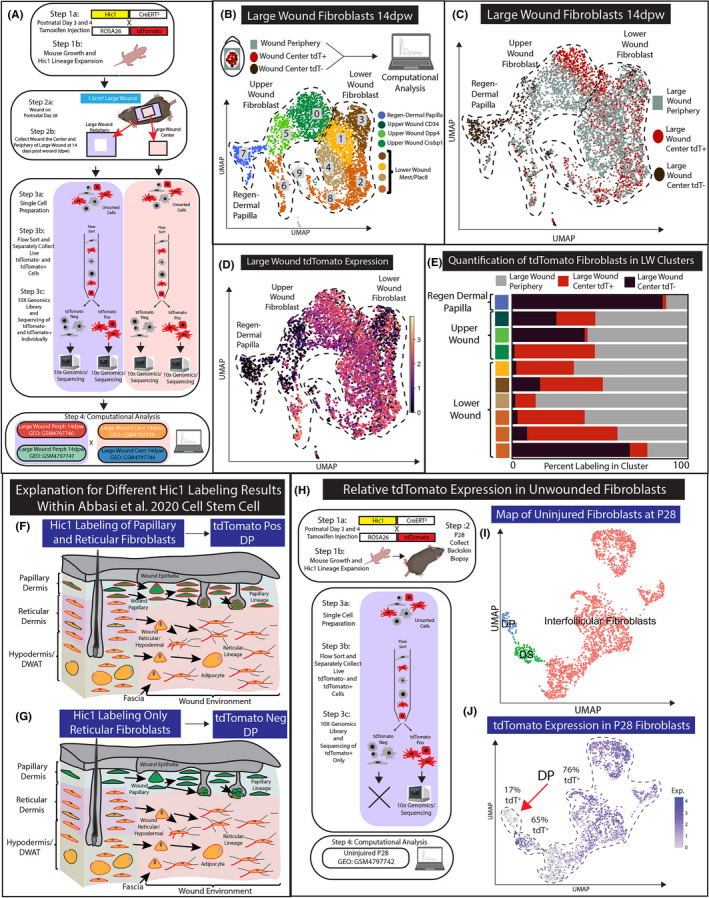
Analysis of Hic1CreERt2‐labelled tdTomato expression in scRNA‐seq data of large wound fibroblast populations. (A) Schematic of experimental design. (B) Recomputed UMAP analysis of all large wound fibroblasts aligned with a genome containing tdTomato. Both wound periphery files (LWP 14dpw Pos and Neg) are coloured grey. (C) UMAP projection colour coded by 10× Genomics libraries of large wounds. (D) UMAP of large wounds with tdTomato expression. (E) Quantification of the number of cells within the Leiden cluster by 10× Genomic library. (F‐G) A proposed explanation for inconsistent results between Hic1CreERT2 histological lineage tracing (F) and scRNA‐seq data (G). (H‐J) An alternative explanation for the lack of tdTomato‐positive cell scRNA‐seq lineage tracing. UMAP projection of fibroblasts subclustered using original Louvain algorithm and grouped as ‘interfollicular fibroblasts’, ‘DP’ or ‘DS’ based on canonical markers (I) (Figure S3). tdTomato expression overlayed on uninjured fibroblast UMAP (J)
